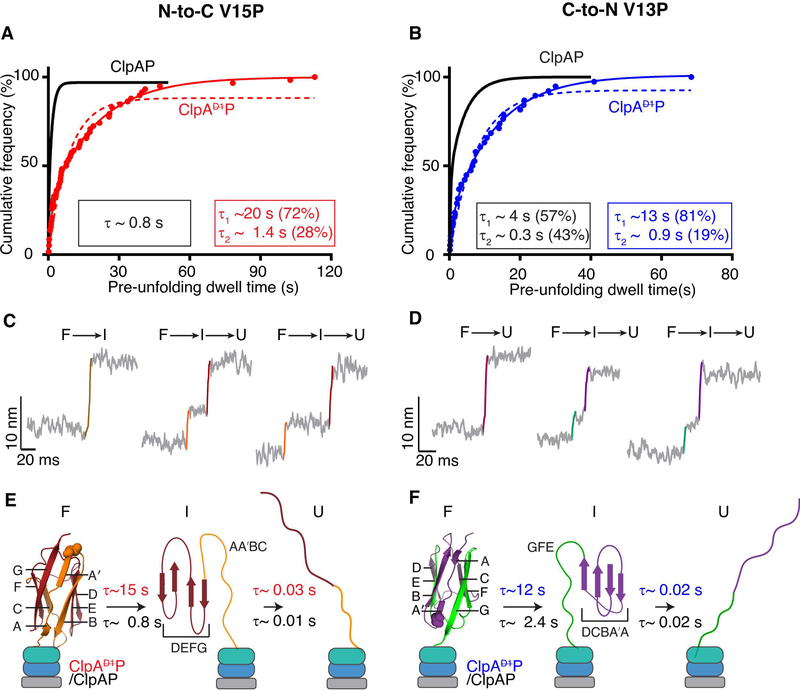
Figure 3: Unfolding by ClpAP and ClpAP.
(A) Cumulative-frequency distribution of pre-unfolding dwell times (n = 58 events) for ClpAP unfolding of V15P domains in the N-to-C direction. The solid red line is a double-exponential fit (R2 > 0.99), with time constants and amplitudes shown in the red rectangle. The red dashed line shows a single-exponential fit. The black line is a single-exponential fit of data for ClpAP unfolding of the same substrate taken from Olivares et al (Olivares et al., 2017; Olivares et al., 2014), with the time constant in the black rectangle. (B) ClpAP and ClpAP unfolding of V13P domains in the C-to-N direction. (For ClpAP:V13P, n = 38 events). Colors and fits are the same as panel (A), except the ClpAP data is a double-exponential fit and ClpAP data are shown in blue. (C, D) Traces, with short-lived intermediates, during the unfolding of titin domains by ClpAP in the N-to-C (C) or C-to-N (D) directions. Raw traces were decimated to 1500 Hz. (E, F) Kinetic and structural models for intermediates formed during enzymatic unfolding of titin domains by ClpAP and ClpAP. β-strands in the titin domain are labeled, and approximate structural elements in the intermediate during unfolding from each terminus are shown. Bootstrap analyses were performed on pre-unfolding dwells reported in A and B and the 95% CI of the distribution of the time constants are reported in Table 1.