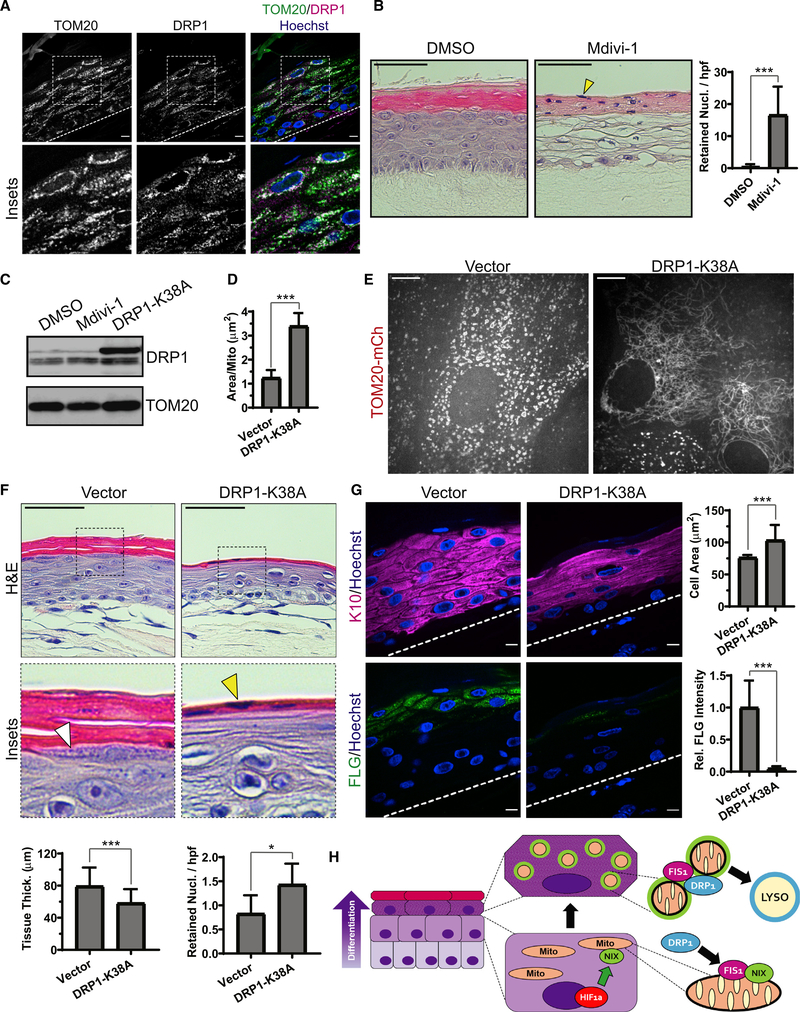
Figure 7. Blockade of mitochondrial fission via DRP1 impairs epidermal differentiation.
(A) IF of DRP1 and TOM20 in mature epidermal cultures.
(B) H&E staining of organotypic epidermis treated with DMSO or Mdivi-1, showing retained nuclei in cornified layers (yellow arrowhead) and (right) quantification of retained nuclei in DMSO- versus Mdivi-1-treated cultures (mean ± SD, n = 70 fields, 4 expts., ***p < 0.0001).
(C) WB of DRP1 in lysates from organotypic cultures treated with Mdivi-1 or transduced with DRP1-K38A.
(D) Quantification of mitochondrial size in upper-layer cells transduced with TOM20-mCh along with DRP1-K38A or a viral vector (mean ± SD, n = 61 fields, 4 expts., ***p < 0.0001).
(E) SDC images of the upper layers of organotypic epidermis expressing TOM20-mCh alone (left) or with DRP1-K38A (right).
(F) H&E staining of cultures expressing a viral vector or DRP1-K38A, highlighting KH granules (white arrowhead) and retained nuclei (yellow arrowhead) and (bottom) quantification of tissue thickness (mean ± SD, n = 103 fields, 4 expts., ***p < 0.0001) and retained nuclei (mean ± SD, n = 101 fields, 4 expts., *p = 0.0115) in vector- versus DRP1-K38A-expressing cultures.
(G) IF of K10 and FLG in organotypic cultures expressing vector or DRP1-K38A and (right) quantification of K10-positive cell size (mean ± SD, n = 30 fields, ***p < 0.0001) and FLG staining (mean SD, n = 30 fields, ***p < 0.0001). Dashed lines mark the bottom of the epidermis. Black scale bars, 50 μm; white scale bars, 10 μm.
(H) Model depicting the onset of NIX expression in differentiating keratinocyte layers driven by hypoxia-regulated signaling; NIX-positive mitochondria (green rim) exhibit enhanced localization of FIS1, which recruits DRP1 to produce mitochondrion fragments that are targeted for lysosomal degradation in the uppermost epidermal layers.