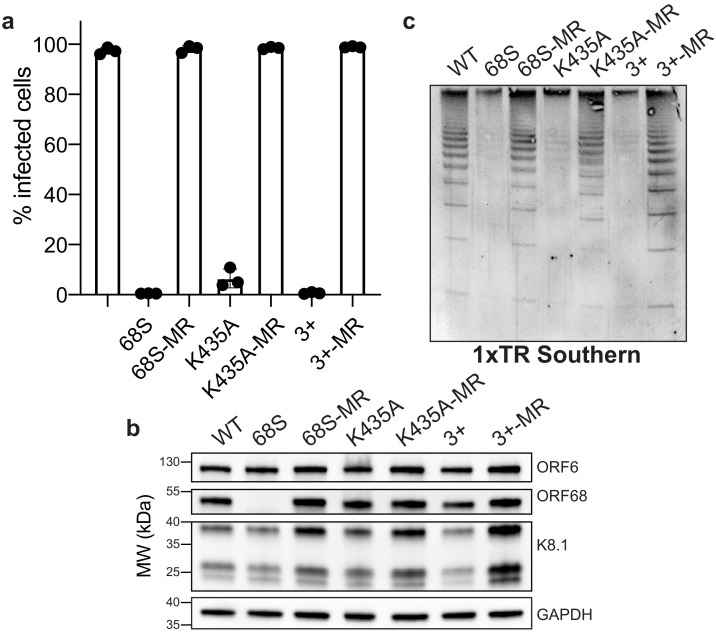
Figure 5. Residues in ORF68 that ablate dsDNA binding in vitro are required for genome cleavage and packaging in vivo.
(a) iSLK cell lines containing ORF68 mutants (68S, K435A, and 3+) and their corresponding mutant rescues (MR) were established using the KSHV BAC16 system. Progeny virion production by these cell lines was assayed by supernatant transfer and flow cytometry of target cells. (b) Western blot of whole cell lysate (25 μg) from ORF68.stop iSLK cell lines. GAPDH was used as a loading control. ORF6 is an early gene and K8.1 is a late gene. (c) Southern blot of DNA isolated from iSLK cell lines using a probe for the terminal repeats. DNA was digested with PstI, which cuts within the genome but not within the terminal repeats and generates a ladder of terminal repeat-containing DNA when successful cleavage and packaging occurs.