Figure 1.
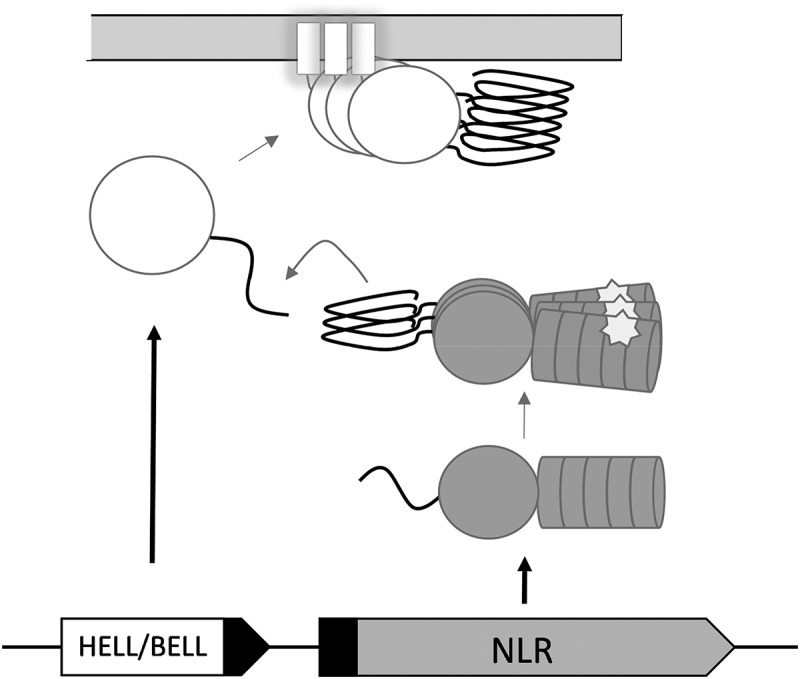
A generic scheme of NLR amyloid signalling
This simplified sketch summarizes key features common to the het-s/nwd2 system of Podospora anserina and to analogous signalosome pathways, described or predicted in other fungal and bacterial species. The signalosome components include an NLR (Nod-like receptor) (for instance NWD2) and a cell-death execution protein (for instance HET-S). The NLR and execution proteins are encoded by adjacent (or even overlapping) genes. The N-terminal part of the NLR and the C-terminal part of the execution protein share a homologous amyloid-forming region (symbolized as a black curved segment). The cell-death execution domain can be of the HeLo or HELL (HeLo-like) type in fungi or BELL type in bacteria (bacterial domain analogous to HELL). The NLR can display a range of architectures such as NACHT/WD, NACHT/ANK or NB-ARC/TPR. Upon binding of a ligand (symbolized as star-shape) in the C-terminal repeat domain, the NLR oligomerizes. This oligomerization induces cooperative folding of the N-terminal motifs into an amyloid fold. This fold is used as template to induce amyloid formation in the homologous region of the execution protein. Amyloid folding triggers activation of the pore-forming activity of the execution protein, which is dependent on the formation of a membrane-targeting α-helix and leads to induction of cell death. Many steps in this pathway remain hypothetical in some or all of the systems described in the text.
