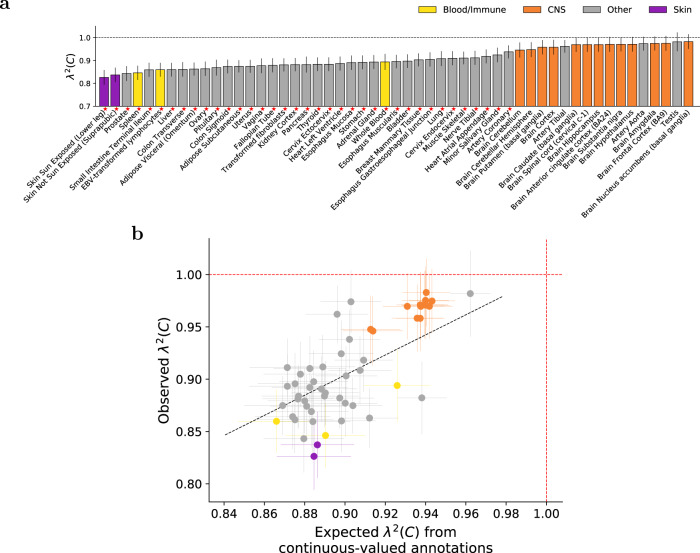
Fig. 4. S-LDXR results for 53 specifically expressed gene (SEG) annotations across 31 diseases and complex traits.
a We report estimates of the enrichment/depletion of squared trans-ethnic genetic correlation (λ2(C)) for each SEG annotation (sorted by λ2(C)). Results are meta-analyzed across 31 diseases and complex traits. Error bars denote ± 1.96 × standard error. P-values are obtained from the standard normal distribution. Red stars (⋆) denote two-tailed p < 0.05/53. Numerical results are reported in Supplementary Data 21. b We report observed λ2(C) vs. expected λ2(C) based on 8 continuous-valued annotations, for each SEG annotation. Results are meta-analyzed across 31 diseases and complex traits. Error bars denote ± 1.96 × standard error of the meta-analyzed λ2(C). Annotations are color-coded as in (a). The dashed black line (slope=0.96) denotes a regression of observed λ(C) − 1 vs. expected λ(C) − 1 with intercept constrained to 0. Numerical results and population-specific heritability enrichment estimates are reported in Supplementary Data 20.