Extended Data Fig. 6. DRM2 interacts with UVR8 and its active form UVR8W285A.
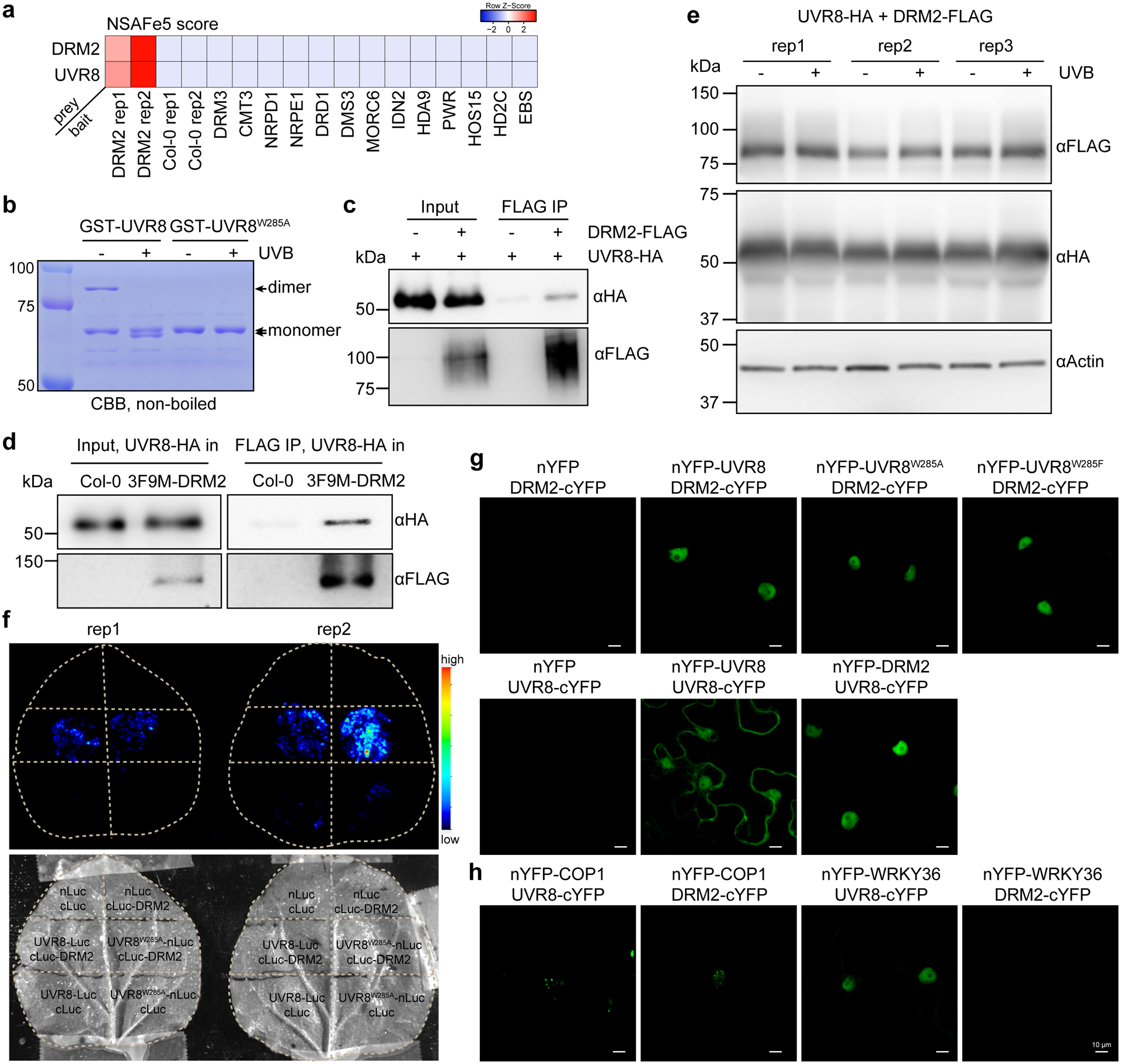
a, Heatmap showing the NSAF score (an indicator of normalized spectral abundance factor) of UVR8 and DRM2 in various immunoprecipitation-mass spectrometry (IP-MS) experiments. The IP-MS data of DRM2 is from ref34, DRM3 and NRPE1 are from ref81, CMT3 is from ref82, DRD1 and DMS3 are from ref83, MORC6 is from ref70, IDN2 is from ref84, HDA9 and PWR are from ref85, HOS15 is from ref86, HD2C is from ref87, EBS is from ref88. b, Coomassie bright blue staining of non-boiled GST-UVR8 proteins on SDS-PAGE. The GST-UVR8W285A proteins serve as control for monomer. c, Co-immunoprecipitation of UVR8 and DRM2 with FLAG beads from N. benthamiana leaves co-expressing UVR8-HA and DRM2-FLAG. d, Co-immunoprecipitation of UVR8 and DRM2 with FLAG beads from transgenic Arabidopsis plants co-expressing UVR8-HA and 3F9M-DRM2. UVR8-HA in Col-0 serves as a control. e, Immunoblots showing protein levels with or without UVB treatment. The ±UVB set-up is the same as that in Fig. 4g. Actin serves as an internal control. f, Split luciferase assay showing the interaction between DRM2 and UVR8W285A. The indicated constructs were co-expressed in N. benthamiana leaves and imaged after spraying with the luciferin. nLuc- and cLuc-only vectors serve as negative controls. Two biological replicates are shown. g, Bimolecular fluorescence complementation (BIFC) assays in N. benthamiana leaves showing the interaction between DRM2 and different forms of UVR8. Scale bar, 10 μm. h, BIFC assays co-expressing indicated proteins in N. benthamiana leaves. Scale bar, 10 μm.
