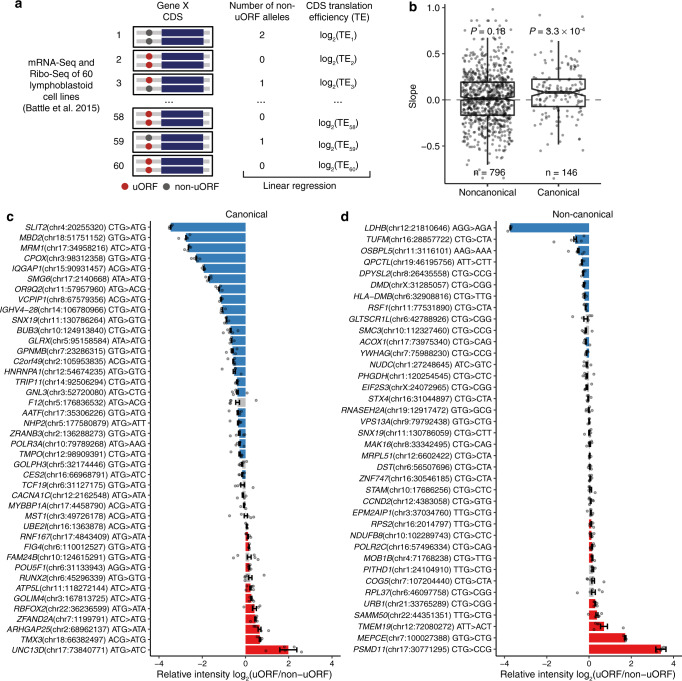
Fig. 7. Experimental verification of canonical and noncanonical upstream open reading frames (uORFs).
a The scheme showing how to determine the effect of uORF variations in the human population on the translation efficiency (TE) of downstream coding sequences (CDSs). With the mRNA-Seq and Ribo-Seq data of 60 human lymphoblastoid cell lines115, we calculated the translation efficiency of CDS for each gene and obtained the genotypes of each subject from the 1000Genomes Project. For each uORF variant, we performed a linear regression between the number of non-uORF alleles and the log2(TE) of downstream CDS across the 60 cell lines. b The distribution of slopes in the linear regression between genotypes (the number of non-uORF alleles) and CDS translational efficiency among 60 cell lines. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 times the interquartile range. The number of variants in each category was shown in the plot. Exact P values of two-sided Wilcoxon signed-rank tests were shown in the plot. The ratio of relative luciferase intensity (log2) between the reporters with the uORF allele or the non-uORF allele for each variant of canonical uORFs (c) or noncanonical uORFs (d). The bars are displayed in blue or red when the relative intensity of uORF-allele is significantly lower or higher than that of the non-uORF allele (one-sided Wilcoxon rank-sum tests, Padj < 0.1), respectively. Measures of center, mean; error bars, standard errors. n = 4 or 5 independent biological replicates for each variant (details are presented in source data). Source data are provided as a Source Data file.