Summary
Inheritance of chromatin-bound proteins theoretically plays a role in the epigenetic transmission of cellular phenotypes. Protein segregation during cell division is however poorly understood. We now describe TrIPP (Tracking the Inheritance Patterns of Proteins): a live cell imaging method for tracking maternal proteins during asymmetric cell divisions of budding yeast. Our analysis of the partitioning pattern of a test set of 18 chromatin-associated proteins reveals that abundant and moderately abundant maternal proteins segregate stochastically and symmetrically between the two cells with the exception of Rxt3p, Fpr4p, and Tup1p, which are preferentially retained in the mother. Low abundance proteins also tend to be retained in the mother cell with the exception of Sir2p and the linker histone H1. Our analysis of chromatin protein behavior in single cells reveals potentially general trends such as coupled protein synthesis and decay and a correlation between protein half-lives and cell-cycle duration.
Subject areas: biological sciences, molecular biology, cell biology
Graphical abstract

Highlights
-
•
Most protein half-lives are equal to cell-cycle length
-
•
Net synthesis rates of 18 test proteins are 2-fold higher than their decay rates
-
•
Most tested proteins segregate stochastically between mothers and daughters
-
•
“Old” Rxt3p, Tup1p, and Fpr4p and low abundance proteins are retained in the mother
Biological sciences; molecular biology; cell biology
Introduction
During asymmetric cell division a stem cell produces two different daughter cells: one cell that preserves the characteristics of “stemness” and will go on to perpetuate the stem cell lineage, and the other that will eventually undergo cellular differentiation into a specific cell type. Since the DNA sequence, barring random mutations introduced by DNA replication is identical in the two daughters, the stem cell phenotype and the differentiating phenotype are both inherited epigenetically: each cell will receive a different set of epigenetically encoded instructions, one to remain a stem cell and the other to start differentiating.
In order to understand how the two phenotypes are established and maintained we need to identify the epigenetic factors that distinguish the two daughters after asymmetric cell division.
Eukaryotic genomes are packaged into chromatin fibers consisting of arrays of nucleosomes: globular histone protein-DNA complexes (Luger et al., 1997). Because chromatin features and chromatin-bound proteins cooperate to regulate transcription, the features that define a specific chromatin state have the potential to transmit epigenetic information about the gene expression state of its underlying locus to the next generation (Petruk et al., 2012).
In order for chromatin features to be truly epigenetic, they have to be accurately “copied” (i.e. re-established on the correct genomic locus) after cell division and they have to be instructive of the transcription state at their genomic location. Because recent studies have provided experimental support for both claims—at least for some chromatin features such as heterochromatic histone marks (Coleman and Struhl, 2017; Laprell et al., 2017; Wang and Moazed, 2017)—it is reasonable to assume that the daughter cell that inherits the maternal phenotype after asymmetric cell division should also inherit those parental chromatin components that presumably define that particular phenotype. Consequently, these parental chromatin components would have to segregate asymmetrically only into one of the two cells, whereas the other daughter should inherit new chromatin components that will commit it to differentiation. On the other hand, chromatin features that are associated with constitutive traits that have to be maintained in both cells would be “inherited” symmetrically in both cells.
It is important to identify which chromatin components carry epigenetic information and establish how these components segregate during genome replication and cellular division, if we want to understand cellular differentiation (pathological or developmental).
Budding yeast cells also divide asymmetrically, resulting in a larger mother and a smaller daughter cell. The mother can generate ∼30 daughters during its replicative lifespan. The mother cell has therefore a phenotypic identity that distinguishes her from her daughters and that determines the length of her replicative lifespan. The aging phenotype of the mother cell has been extensively studied, and several models have been proposed to explain asymmetric segregation of aging factors. Yet epigenetic factors that define the “mother” and “daughter” phenotypes and the role of chromatin in determining those phenotypes are still largely unknown. According to currently proposed models, the aging phenotype is caused by molecular “aging factors.” An aging factor has to satisfy three criteria: it has to accumulate over time, it has to be preferentially retained in the mother cell, and it has to directly or indirectly lead to cell death. Several candidates for “aging factors” have been proposed, the most attractive being the following: (1) extra chromosomal ribosomal DNA circles (ERCs) (Denoth-Lippuner et al., 2014), (2) protein carbonyls, and (3) proteins damaged by oxidation and old mitochondria (reviewed in (Steinkraus et al., 2008)). Because none of these factors completely satisfy all the criteria, a consensus on the determining aging agent has still not been reached, and epigenetic factors that define the “mother” phenotype and the role of chromatin in the process are still not known.
Several studies in recent years have identified proteins that accumulate asymmetrically in only one of the two cells after division. Yang et al. (Yang et al., 2015) found 74 mother-enriched and 60 daughter-enriched yeast proteins; however, their assay could not differentiate between maternal proteins that potentially carry epigenetic information and newly synthesized copies. It is therefore unclear whether any epigenetic information is transmitted by chromatin-associated proteins, which incidentally had a tendency to accumulate in the daughter cell in their assay. On the other hand, Thayer et al. (Thayer et al., 2014) looked specifically for long-lived maternal proteins that accumulate in the aging yeast mother cell. They mostly identified cytoplasmic and membrane proteins however, which probably do not play a role in the epigenetic inheritance of specific gene expression states even though they could potentially contribute to the aging phenotype of the mother cell, although the latter has not been explicitly tested. Finally, Garcıá del Arco et al. (Garcia Del Arco et al., 2018) show that during cell division of the fly midgut epithelium, the maternal CENP-A centromeric histone variant segregates asymmetrically into the daughter cell that will remain a stem cell.
In order to find chromatin components that segregate asymmetrically during yeast cell divisions, we need to be able to follow the partitioning of maternal proteins for several generations in single-cell lineages. Most methods currently used to monitor the dynamics of the cellular proteome, including the ones used in the studies mentioned earlier, are based on cell populations and combine mass spectrometry or imaging for proteome analysis with cytometry or biochemical methods to separate mother and daughter cell populations and do not provide information on protein dynamics in single cells.
We therefore set out to develop a live cell imaging screen aimed at identifying chromatin factors that could potentially be involved in epigenetic inheritance of cellular phenotypes during asymmetric cell division. We used the photo-convertible fluorescent protein Dendra2. Because photo-conversion from green to red fluorescence after UV light irradiation is irreversible, one can directly measure the half-lives of maternal Dendra2 fusion proteins in each cell. Maternal proteins can be followed for several cell generations (one yeast cell generation is typically 90–100min long), unlike with a similar system using a tandem sfGFP-mCherry cassette that can track newly synthesized proteins within a window of 45 min from synthesis because of the difference in folding kinetics of the fast-folding sfGFP, which starts fluorescing within minutes from synthesis, and the slow-folding mCherry, which takes ∼45 min to mature (Khmelinskii et al., 2012).
Because the UV pulse for photo conversion lasts only 1min, we can start to track maternal proteins almost immediately and more importantly we can do it during the first cell cycle and after the first cell division following photo conversion. This would not be possible with the other imaging technique developed to detect “old” proteins that is based on SNAP/CLIP tagging of proteins of interest. Labeling of SNAP/CLIP-tagged proteins requires electroporation of fluorescent dyes into yeast and a 2-h pulse step followed by a chase step (Stagge et al., 2013). Given that the budding yeast cell cycle typically lasts from 1.5 to 2 h, yeast would have undergone at least one cell division during the labeling pulse and we would have been able to follow only longer lived abundant proteins starting from the second division after fluorescent labeling.
We used TrIPP to monitor cell cycle dynamics of a test set of 18 proteins. Remarkably, our analysis of this small set of proteins has revealed a pattern for protein dynamics during the cell cycle that may turn out to be more general: moderately and highly abundant maternal chromatin proteins (more than ∼3000 molecules/cell) segregate stochastically between the mother and daughter cells with a cell population mean for the fraction of proteins retained in the mother at ∼0.5 as expected, whereas low abundance proteins (between ∼600 and ∼3000 molecules/cell) are mostly retained in the mother. There were, however, some exceptions: moderately abundant metabolic gene regulators Rxt3p (a subunit of the histone deacetylase complex Rpd3L), Fpr4p (proline isomerase of H3P38), and Tup1p (transcription repressor) are preferentially retained in the mother cell and low abundance proteins Sir2p (part of the heterochromatic complex Sir) and H1 (linker histone) segregate stochastically like more abundant proteins.
Results
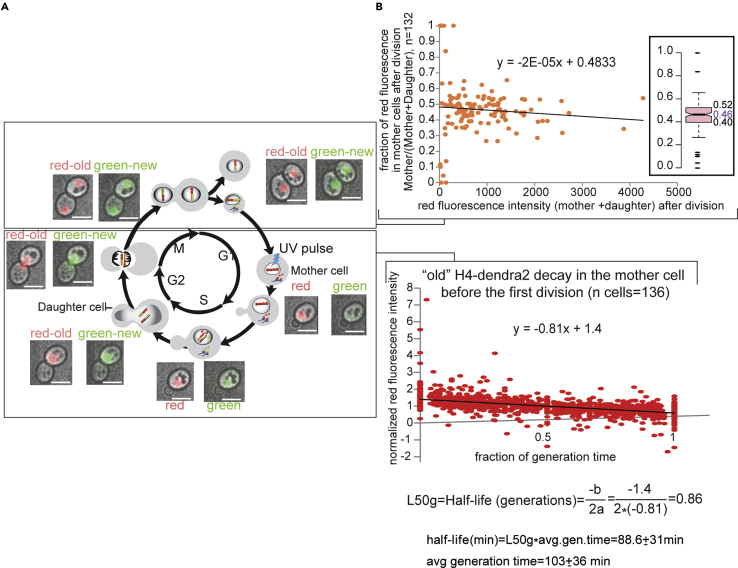
As proof of principle for TrIPP, we have constructed a test set of 18 strains carrying chromatin-associated proteins (including chromatin remodelers, histones, histone modifiers, and heterochromatic proteins) fused to the photoconvertible fluorescent protein Dendra2 (Table S1)). The set of 18 proteins comes from a larger set of ∼60 Dendra2 fusion strains and represents fusion proteins with a sufficiently high signal-to-background ratio suitable for quantitative analysis. Dendra2 switches irreversibly from green to red fluorescence after UV/blue light irradiation. Consequently, live fluorescence imaging ([HiLo] ,(Lim et al., 2011)) of dividing cells allows us to distinguish between already synthesized maternal proteins, which emit in the red spectrum after a UV pulse, and newly synthesized green proteins made after the pulse (Figure 1).
Figure 1.
Tracking segregating maternal chromatic proteins
(A) Chromatin proteins are fused to the photo-convertible fluorescent protein Dendra2. Proteins already incorporated in chromatin will switch from green to red fluorescence after UV light irradiation. Newly synthesized green fluorescent fusion proteins are incorporated into daughter chromatids after the UV pulse, and segregation of “old” red and “new” green proteins is monitored by live cell imaging.
(B–D) HiLo imaging of Spc42p (B) (component of SPB-Spindle Pole Body) fused to Dendra2. The “old” (red) SPB segregates into the daughter cell, as previously reported. Histone H2B (C) segregates symmetrically but can also be preferentially retained in the mother cell: compare red signals between M3D1 and its daughter at the 240.5 min time point. Maternal Rxt3p (subunit of the histone deacetylase Rpd3L) is retained in the mother cell (D). Cell lineages are shown below the images.
See also Videos S3A, S3B, and S3C for an example of Rxt3p segregation.
We can therefore record segregation patterns of maternal chromatin proteins and follow cellular localization dynamics of fusion constructs through mitosis. Due to closed mitosis in yeast, nuclear localization during mitosis does not automatically signify chromatin association. Nevertheless, the inheritance of maternal proteins within the nucleus—as is the case for all tested proteins—is still an indication that these chromatin proteins may remain associated with mitotic chromosomes and could therefore be epigenetically inherited (for examples see Figures 1C and 1D and Videos S1A, S1B, S1C, S2A, S2B, S2C, S3A, S3B, S3C, S4A, S4B, and S4C).
We used Spc42p-dendra2 as a positive control. Spc42p is part of the spindle pole body (SPB). Our analysis shows a preferential inheritance of the “old” SPB by the daughter cell as previously reported (Hotz et al., 2012) (Khmelinskii et al., 2012) (Figure 1B). Our conditions for UV irradiation do not affect cell growth and viability as doubling times, and the numbers of dividing cells are comparable between non-irradiated cells or cells grown in liquid media and UV irradiated cells (compare Figures 3B and S4B).
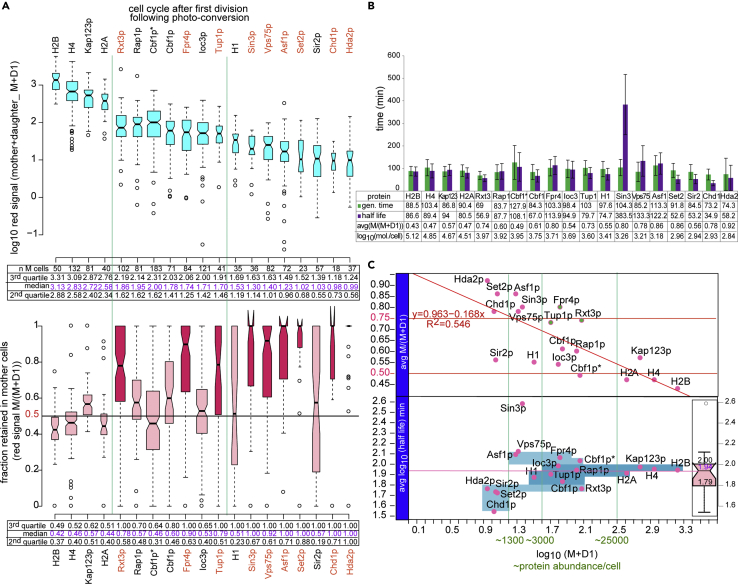
Figure 3.
Segregation of 18 maternal chromatin proteins between mother and daughter cells after mitosis
(A) Box plot distributions of average red fluorescence intensities (calculated as described in Figure 2B) from the mother M and her first daughter cell D1 after photo-conversion (the number of analyzed mother/daughter pairs is indicated in the table below the top graph) (top) and the fraction of the total red fluorescence intensity shown on top that is retained in the mother M/(M + D1) (bottom). The tables below the graphs show the median values of the entire distribution (magenta) and the medians of the second and third quartile for each dendra2 protein fusion. The M and D1 values recorded for Ioc3p and Sir2p have been divided by 2 to correct for the double dendra2 tag of these two constructs. Cbf1p∗ is from the strain with a lysΔ background that was used in the half-life measurements by mass spectrometry (Figure S4). See also Figure S2 for scatterplots of mother retention versus protein abundance per cell for all 18 test proteins.
(B) Bar graph showing average cell generation times and half-lives of indicated dendra2 fusion proteins. Generation times were averaged over 2–3 generations for each mother cell (number of cells indicated in A). Half-lives were determined from the decay of red fluorescence during the first cell cycle after photo-conversion in mother cells and calculated as fractions of generation time as described in Figure 2B and then converted to minutes using the average generation time shown in the graph. The error bars represent the standard deviation from the average generation time. Maternal protein abundance (PA, sea-green x axis in (C) is proportional to the average total red fluorescence in the mother and daughter cells after mitosis (M + D1) for all measured mother-daughter pairs: PA = a∗(M + D1). a = 70,000/((M + D1) of H4)). 70,000 is the estimated number of dendra2-histone fusion proteins bound to chromatin per cell. Because only one histone gene out of the two copies in the yeast genome has the dendra2 fusion, the number of histones with the dendra2 tag is equal to the number of nucleosomes in the cell: . The efficiency of photo-conversion (ranging from 50% to 90%, data not shown) has not been taken into account in the calculation because conversion efficiencies varied from cell to cell and only an estimate of the order of magnitude of protein abundance is sufficient for the analysis.
(C) Correlation between protein abundance and half-life (bottom panel) and retention in the mother cell after mitosis (top panel). Bottom panel: the four blue rectangles (bottom to top) encompass proteins from the first to the fourth quartile from the box plot distribution of half-lives on the right, respectively. Low abundance proteins (<1300/cell) tend to be in the first quartile and high and moderately abundant proteins (>3000/cell) tend to be in the third and fourth quartiles, with the exception of Sin3p (a low abundance protein with a long half-life). Top panel: highly (>25000/cell) and moderately (between 3000 and 10000/cell) abundant maternal proteins are distributed stochastically between the mother and the daughter during mitosis, with a mean retention in the mother around 50% and variance inversely proportional to protein abundance (as shown in A). Rxt3p, Fpr4p, and Tup1p with a high retention bias in the mother are the exception. Low abundance proteins (between 600 and 3000/cell) have a clear retention bias for the mother cell with the exception of H1 and Sir2p, which are distributed stochastically similar to highly abundant proteins. See also Videos S1A, S1B, S1C, S2A, S2B, S2C, S3A, S3B, S3C, S4A, S4B, and S4C for examples of protein segregation and Figures S4 and S6 and Table S2 for comparison to half-life values obtained by mass spectrometry.
Half-life measurement and protein segregation after cell division
The segregation patterns of maternal chromatin proteins between mother and daughter cells are estimated from the fraction of maternal proteins remaining in the mother cell after the first cell division following photo conversion (Figure 2).
Figure 2.
Determination of the half-life and maternal protein segregation pattern for H4-Dendra2
(A) Old-red and new-green histone H4 distribution in the mother and daughter cells during the yeast cell cycle followed by HiLo live fluorescence microscopy. Cells were illuminated with UV light in G1, and images in bright field and the red and green channels were taken every 6.5 min for 6 h. The micrographs show a representative mother cell at indicated stages during the first cell cycle after photo-conversion. Images from the green and the red channels were each merged with the corresponding bright field image. A 5-μm white scale bar is shown in the bottom right corner of each micrograph. See also Videos S2A, S2B, and S2C for an example of H4 segregation.
(B) Half-life (bottom) and old/maternal H4-dendra2 partition (top) between mother and daughter cells. The x axes represent all the time points taken before the first cell division after photo conversion from 136 mother cells calculated as fractions of generation time for each mother cell (bottom) and the sum of average red fluorescence intensities in the mother and her first daughter produced after photo conversion over the duration of the mother's subsequent cell cycle (i.e. before the production of the second daughter after photo conversion) (top). The average generation time is an average of up to 3 cell-cycle lengths from 136 mother cells. The red fluorescence intensities in both graphs have been subtracted from background fluorescence. The signal in the Y axis of the bottom graph has also been normalized to the average fluorescence intensity from the time of photo conversion to the time of the first cell division for each mother cell. The average fraction of maternal H4 retained in the mother cell after cell division is estimated from the Y axis cutoff in the top graph (0.48) and the median value in the box plot (top inset) (0.46). See also Figure S1 for photo bleaching test.
The decrease in the amount of maternal proteins in the mother cell over time is determined from the decay rate of red fluorescence in the mother cell after photo conversion and before the first cell division. The measured rate of decay is used to calculate the half-life of the dendra2 fusion protein. Photo-bleaching did not affect the decay rate (Figure S1). The half-life of red fluorescence was calculated as described in Figure 2B and in Transparent methods in Supplemental information. The currently used methods for measuring protein half-lives are based on pulse-chase time-courses with labeled amino-acids in cell populations and require either careful calibration of total protein content between different time points or theoretical approximations of protein synthesis rates as in the classic [35S]-methionine and [35S]-cysteine pulse chase experiments (Kornitzer, 2002) or SiLAC MS experiments (Schwanhausser et al., 2011, 2013), respectively. These experimental drawbacks are circumvented with our method because it is based on observations of single cells in vivo and it allows for direct and independent measurements of protein decay and net synthesis rates in each cell.
Low abundance proteins and metabolic gene regulators Rpd3L, Fpr4p, and Tup1p are retained in the mother
Half-lives and segregation patterns of maternal proteins for all 18 dendra2 fusions are summarized in Figure 3. Our analysis reveals an inverse correlation between protein retention in the mother cell and protein abundance. If protein partitioning between the mother and the daughter were purely stochastic, we expect a normal distribution of protein fractions retained in the mother with a mean ∼0.5 and a variance that is inversely proportional to protein abundance. Indeed, we observe that highly abundant proteins (estimated at >25000/cell, see Transparent Methods in Supplemental Information) are divided between the mother and the daughter into two equal sets in most cells as shown for histone proteins and Kap123p (nuclear transporter of histone proteins). Moderately abundant proteins (between 3000 and 6000 per cell) also segregate equally on average in the cell population but with bigger variability between cells, i.e. there is a higher fraction of mother or daughter cells in the population that have inherited disproportionally more maternal proteins. Moreover, if protein partitioning is random, the cells that inherit more proteins should be mothers or daughters in equal proportions, as we see for Rap1p (general transcription factor also involved in heterochromatin formation), Cbf1p (associates with centromeric DNA elements and kinetochore proteins), and Ioc3p (subunit of the nucleosome remodeler Isw1a).
Finally, following the same reasoning, small quantities of proteins (less than 1300/cell) will segregate asymmetrically in an even larger fraction of cells, ending up in mothers and daughters in equal proportions, than moderately abundant proteins, but the mean of the protein fraction retained in the mother should still be ∼0.5. Surprisingly, this behavior is observed only for Sir2p (subunit of the heterochromatic Sir complex) and the linker histone H1. The other low abundance proteins Sin3p, Vps75p, Chd1p, Asf1p, Set2p, and Hda2p are all preferentially retained in the mother, as are moderately abundant proteins: Rxt3p, Fpr4p, and Tup1p (Figures 3A and 3C). Due to the stochastic nature of protein distribution between the mother and the daughter cell, highly and moderately abundant proteins in cells that had a low red signal because of incomplete photo conversion also partition asymmetrically but with a similar bias towards the mother or the daughter cell (see H4, Cbf1p, Rap1p and Ioc3p in Figure S2). This argues against the possibility that the observed preferential retention of low abundance proteins in the mother cell is due to an imaging artefact that favors detection of low signal in the mother cell, because we repeatedly record asymmetric segregation of weak red fluorescence into daughter and mother cells in equal proportions for all proteins that segregate stochastically. In addition, the red signal intensities M and D (for mother and daughter cell, respectively) used for the calculation of the M/(M+D) ratio represent the mean signal over the whole length of the cell cycle. Consequently, any potential bias toward the mother cell that could have been due to the initial size difference between the mother and the daughter would be negligible because the daughter would have reached the mother cell’s size in the first part of the cycle. The size difference between the mother and the daughter is also unlikely to be a significant source of red signal bias toward the mother because the signal is localized to the nucleus, which is of comparable size in the mother and the daughter cell almost immediately after division.
Half-life durations of abundant proteins are equal to cell-cycle length
Our measurements of decay rates of red fluorescence also revealed that the half-lives of the examined proteins are comparable to cellular generation times except for low abundance proteins whose half-lives are ∼50% of generation time (Figures 3B and 3C). This means that highly and moderately abundant proteins turn over completely in two generations. These proteins could therefore potentially transmit epigenetic information from one cell generation to the next if they stay associated with the relevant genomic loci after DNA replication and mitosis. The case for epigenetic inheritance of low abundance proteins is somewhat more difficult to make. Because these proteins turnover within one cell generation, epigenetic information could only be transmitted if newly synthesized proteins were exchanged with the old proteins directly on chromatin in order to preserve and transmit the information on their underlying genomic locations.
Curiously, Sin3p (component of Rpd3S/L histone deacetylase complexes) appears to have a half-life that is almost four times longer than the cell generation time. Although the significance of this result is not clear, it is also the only half-life value that matches half-life estimates measured by pulse SiLAC MS (Christiano et al., 2014). The other half-lives are on average six times shorter than the MS measurements from Christiano et al. (Christiano et al., 2014) (Figure S4). One possible explanation for this discrepancy is that the dendra2 tag destabilizes the protein. Our results from a mass spectrometry SiLAC assay of cell extracts from two dendra2 strains (CBF1dendra2 and H1dendra2) from a time course after switch from heavy lysine to light lysine (Figure S4B and Table S2) show that the tagged and untagged proteins have similar half-lives, suggesting that dendra2 has no effect on protein stability. Moreover, our half-life estimates from the SiLAC experiment are closer to our half-life measurements from dendra2 fluorescence decay (Figure S4C) than to the published values mentioned earlier (Christiano et al., 2014). The discrepancy is therefore not due to the technical differences or experimental conditions in either experimental method used to measure half-lives.
The difference stems instead from the calculations used to derive half-lives in (Christiano et al., 2014) and our own calculations. Unlike in our assay where the protein decay rate is measured directly from the decay rate of red fluorescence in each cell, in SILAC-LC-MS/MS the protein decay rate is indirectly estimated from the decrease in the H/L ratios (Figure S6 and Transparent methods in Supplemental information) in a cell population over time after the switch to “light” medium. It is consequently necessary to make several assumptions in order to estimate protein decay rates and half-lives. First, it is generally assumed that protein decay is an exponential process (Christiano et al., 2014; Schwanhausser et al., 2011), although that seems not to be the case for some proteins (McShane et al., 2016). Second, protein synthesis rates also need to be estimated because they are not measured in the MS assay.
The calculations in (Christiano et al., 2014) were based on half-life measurements in mammalian cells from (Schwanhausser et al., 2011) where it was assumed that total protein amounts double every cell generation. This assumption has been confirmed for 40 proteins in mouse ES cells (Alber et al., 2018) but does not seem to hold for yeast. According to our direct measurements of net protein accumulation rates described earlier, the cellular content for 13 out of the 18 proteins we measured does not double within one cell cycle (Figure S5C). As a consequence of that assumption, half-lives of proteins with actual synthesis rates that are higher than the 2-fold increase in one cell generation will be underestimated, whereas the half-lives of proteins with low synthesis rates will be overestimated as shown in Figure S4C. In order to be able to compare our half-life values with the published dataset, we nevertheless used the same approximations in our calculations. Surprisingly, we obtain values that are at odds with the ones in (Christiano et al., 2014). The major difference in our calculations and the ones in (Christiano et al., 2014) is in the use of a correction for protein dilution due to cell division applied in the latter. Maternal protein amounts are reduced 2-fold every cell generation because ∼50% are passed on to the daughter in most cases (Figure 3), but because we use total cell extracts from a population of mothers and daughters in the SILAC experiment the only process that causes the decrease in “old/heavy” proteins in the cell population is protein degradation. The correction used in Christiano et al. (2014) (Christiano et al., 2014) is therefore not necessary and is actually the cause for the overestimation of protein half-lives in that study.
Tup1p that was synthesized before S-phase is retained in the mother cell
Tup1p is mostly a general transcriptional repressor that shuts down transcription of >150 genes in response to various environmental signals (reviewed in (Smith and Johnson, 2000)), although it can also act as a transcriptional activator in certain conditions (Proft and Struhl, 2002). A subset of its targets is involved in glucose or galactose metabolism, and Tup1p is necessary for long-term transcriptional memory and faster reactivation of galactose inducible GAL genes (Sood et al., 2017). We consequently wanted to explore further the potential links between Tup1p retention in the mother cell and its function as a repressor of glucose and galactose metabolism genes.
We first checked the repression activity of Tup1p-dendra2 in galactose and dextrose (Figure S3A and Table S3). Although Tup1p-dendra2 represses either glucose metabolism genes in galactose medium or galactose metabolism genes in glucose medium, as expected, the activity of the tagged protein is not as efficient as that of the untagged protein. The efficiency of repression in the TUP1-Dendra2 strain is at an intermediate level between the wt strain and the tup1 deletion strain, most likely because tup1 mRNA levels are reduced in the TUP1-Dendra2 strain (Figure S3A). We thereby conclude that the Tup1p-dendra2 protein is structurally functional but that the Tup1p-dendra2 strain displays a hypomorph phenotype due to reduced Tup1p levels.
Because cells were grown in glucose for the original assay described earlier and considering that Tup1p targets different sets of genes in different carbon sources, we decided to test whether Tup1p is also retained in the mother cell when grown in galactose or when carbon sources are switched. TUP1-Dendra2 cells were grown in galactose or glucose throughout the assay or switched from dextrose to galactose and vice versa before imaging (Figure 4A). The density plot distributions of Tup1p segregation bias (the fraction of red signal retained in the mother) shown in Figure 4A display a bimodal distribution with one subpopulation in which the “old” photo-converted Tup1p-Dendra2 stays in the mother cell and the other in which the photo-converted Tup1p is equally partitioned between the mother and the daughter. Interestingly, the timing of photo-conversion relative to the cell cycle determines if the population will skew more towards mother retention or towards equal Tup1p partitioning.
Figure 4.
Tup1p molecules present in the nucleus before replication are retained in the mother cell
(A) Bean plot distribution of the fraction of maternal (”red”) Tup1p retained in the mother (M/(M + D)) in indicated growth conditions (illustrated in the diagrams above the bean plots). Black lines represent individual mother cells, magenta lines represent multiple mother cells with the same M/(M + D) value. Cells were divided into two populations: mother cells in which the time since the UV pulse for photo-conversion was longer (photo conversion early in the cycle, orange density plot) or shorter (photo conversion late in the cycle, blue plot) than 0.55∗(avg. cell cycle length).
(B) Model for “old” Tup1p retention in the mother cell. Tup1p is shown tethered to the Nuclear Pore Complex as an example for illustration purposes. It is equally possible that Tup1p is directly or indirectly anchored to another nuclear structure such as the nuclear matrix or the nuclear envelope. See also Figure S3 for RT-qPCR of Tup1p target genes and cytometric measurements of G1 length in carbon source conditions from (A).
The timing of photo conversion is estimated from the length of time elapsed between the UV pulse and the time of complete separation of mother and daughter nuclei in the first division after photo conversion for each mother cell. We propose the following hypothesis to explain why Tup1p is preferentially retained in the mother only when photo conversion happens early in the cell cycle. We propose that when photo-conversion happens in G1 before replication (early photo conversion) the red signal is retained in the mother and when photo conversion happens after replication (late photo conversion) the red signal partitions equally between the mother and the daughter. After replication, both newly synthesized Tup1p and old Tup1p (that was present in the nucleus before replication) bind to replicated gene targets, and the gene copies bound with Tup1p are divided equally between the mother and the daughter. If photo conversion happens before replication only one gene copy will carry an old “red” Tup1p molecule after replication and the other will be bound by a new “green” Tup1p protein. The “red” copy will stay in the mother and the “green” copy will go to the daughter. On the other hand, if photo conversion happens after replication, both replicated gene copies will be bound by “red” Tup1p proteins and the red signal will be partitioned equally between the mother and the daughter after division. The probability that photo conversion will happen before replication increases with the length of the G1 phase, whose duration varies depending on culture conditions (Figures S3B–S3D). If G1 is short, early photo conversion would “catch” Tup1p before replication in some cells and after replication in others, resulting in a bimodal distribution of M/M+D with peaks at 1 and 0.5, as observed when cells are grown in glucose or switched from glucose to galactose. Late photo conversion in these conditions happens mostly after replication, resulting in a normal distribution of M/M+D with a mean of 0.5. Conversely, if G1 is of medium length, early and late photo conversion happen mostly before or after replication, respectively, resulting in normal distributions of M/M+D with a mean of 1 or 0.5, respectively. This is observed when cells are grown in galactose. On the other hand, the transition from galactose to glucose causes a lengthening of the G1 phase, resulting in preferential mother retention in a majority of cells because most cells in the population were pulsed with UV before replication even when photo conversion happened later in the cycle.
We now propose a model for the mechanism of Tup1p retention in the mother cell. Tup1p is directly or indirectly anchored/tethered to a nuclear structure (such as the nuclear matrix or the nuclear envelope) during the G1 phase as illustrated in Figure 4B. Because—per our model—tethering occurs exclusively in G1, the newly synthesized Tup1p is “free” to move into the daughter cell where it will be tethered to the daughter’s nucleus during the daughter’s first G1 phase. Meanwhile the gene copy bound with the “old” tethered Tup1p will remain in the mother cell.
Our model for the timing of Tup1p anchoring that would be restricted to G1 is supported by recent results from Sugiyama and Tanaka (Sugiyama and Tanaka, 2019) that show symmetric segregation of Tup1p that has been synthesized in G1 and at the beginning of S-phase. Our model predicts that Tup1p that was synthesized after G1 (i.e. in S-phase and G2) would not be anchored to the nucleus and will eventually outnumber the tethered Tup1p due to constant protein degradation and synthesis throughout the cycle. Total Tup1p including the fractions synthesized in G1 (“tethered” and staying in the mother) and at the beginning of S-phase (“untethered” and segregating stochastically between the mother and the daughter) will therefore appear to segregate symmetrically as has been observed in the study mentioned earlier and our experiments.
Net protein synthesis is 2-fold faster than protein degradation
Because the intensity of green fluorescence after photo conversion is proportional to the amount of newly synthesized protein, we were able to derive protein synthesis rates from the rates of increase in green fluorescence during the first cycle after photo conversion (Figure S5 and Trasparent methods in Supplemental information). The green fluorescence signal was not subject to photo bleaching as shown in Figure S1B. We also determined the distribution of green fluorescence in the mother and the daughter after cell division as we have done for red fluorescence. The fraction of green fluorescence in the mother cell is however less informative about protein inheritance patterns, because the amount of green proteins in the daughter is the sum of proteins inherited from the mother and new proteins that are synthesized in the daughter. Interestingly, even with this limitation, we can still detect the preferential retention of Rxt3p and low abundance proteins Set2p, Hda2p, Asf1p, and Vps75p in the mother cell (Figure 5A). Thus, the tendency of low abundance proteins to stay in the mother cell is also evident in the “green” protein population (Figure 5B) that has not undergone photo-conversion, suggesting that the segregation pattern observed for “red” proteins is not due to damage potentially caused by the UV pulse.
Figure 5.
Segregation patterns between mother and daughter cells after mitosis and synthesis rates of 18 chromatin proteins
(A) Box plot distributions of average green fluorescence intensities (calculated as described in Figure S5B) from the mother M and her first daughter cell D1 after photo-conversion (the number of analyzed mother/daughter pairs is indicated in the table below the top graph) (top) and the fraction of the total green fluorescence intensity shown on top that is retained in the mother M/(M + D1) (bottom). The tables below the graphs show the median values of the entire distribution (magenta) and the medians of the second and third quartile for each dendra2 protein fusion. The measured (M + D) values for Ioc3p and Sir2p have been divided by 2 in the top graph to correct for the double dendra2 tag of these two constructs. Cbf1p∗ is the strain with a lysΔ background that was used in the half-life measurements by mass spectrometry (Figure S4).
(B) Correlation between new (green) protein abundance per cell and green protein retention in the mother cell. The number of molecules per cell was estimated from the total green fluorescence in the mother and daughter cells after mitosis (M + D1): PA = a∗(M + D1). a = 70,000/((M + D1), H4). 70,000 is the estimated number of dendra2-histone fusion proteins bound to chromatin per cell calculated as in Figure 2B. Because most of these proteins are continuously synthesized, we cannot distinguish between green proteins that were inherited from the mother and the ones that were newly synthesized in the daughter cell. Consequently, most green proteins are equally distributed between the mother and the daughter except for Asf1p, Hda2p, Set2p, and Rxt3p thath are retained in the mother as observed for maternal (old) proteins (Figure 3). The low abundance proteins have a tendency to stay in the mother cell as observed for “old” maternal proteins.
(C) Bar graph showing average net protein accumulation rates (Am), protein decays rates (Dm), and protein synthesis rates (Sm) in molecules/min calculated as described in Figure S5B. Am, Dm, and Sm rates were converted from fold change/min (see Figure S5B) to molecules/min by multiplying each rate with the corresponding PA (protein abundance per cell calculated as in B).
(D) Correlation between protein accumulation rates (Am) and protein decay rates in molecules/min. Values are based on green fluorescence intensity as in (B and C). Synthesis and decay rates appear to be co-regulated with decay rates ~2-fold slower than protein accumulation rates (net protein production) (1/0.498 = 2.01), with the exception of Sin3p (net synthesis 13-fold faster than decay) and Set2p (decay is 8-fold faster than net synthesis).
(E) Correlation between old (red) and new (green) protein abundance (PA) per cell (log 10(protein molecules/cell)) calculated as in (B). Due to faster synthesis rates compared with decay rates there is consistently more new “green” protein than old “red” protein in the cell. Note that the equal numbers of old and new histone is a consequence of normalization of all fluorescence intensities to the fluorescence intensity of H4 and the assumption that nucleosomes contain equal amounts of old and new histones as explained in (B). See also Figure S5 for calculations of synthesis rates.
Our independent measurements of protein synthesis (using the population of “green” proteins) and decay (using the population of “red” proteins) have revealed an unexpected coupling of the two processes (Figure 5D). Surprisingly, protein degradation is correlated with protein synthesis with a net protein accumulation rate at ∼2-fold the decay rate for most proteins we tested.
Discussion
We have developed TrIPP—a live cell imaging method for quantitative measurements of protein behavior in single cells that allowed us to directly measure degradation and net synthesis rates as well as inheritance patterns of 18 chromatin proteins. Our method does not perturb the progression of the cell cycle because the dendra2 tag and the UV pulse for photo-conversion do not significantly affect protein stability as shown by similar protein half-lives measured by microscopy and SILAC-LC-MS/MS.
Our results suggest that “old” low abundance proteins have a tendency to remain in the mother cell after mitosis. The biological significance and the generality of this observation still remains to be confirmed on a larger set of proteins. We speculate, however, that the retention in the mother cell may be related to the potentially higher levels of oxidative damage in these proteins, which may also be the reason for their shorter half-lives. These proteins may have reached the end of their useful lives and are not being transferred to the daughter cell because their function has completely deteriorated by the time of cell division.
The retention of maternal Fpr4p, Rxt3p, and Tup1p in the mother cell is especially interesting, because all three are moderately abundant proteins that have been implicated in the regulation of transcription of inducible genes from metabolic pathways (Nelson et al., 2006; Lee et al., 2018; Sood et al., 2017). Fpr4p is a proline isomerase specific for P38 on histone H3. The activity of Fpr4p inhibits methylation of H3K36 at promoters, which stimulates the induction of MET16 and HIS4 (Nelson et al., 2006). Tup1p and Rxt3p have both been implicated in the transcriptional memory of genes that are induced or repressed during carbon source shifts, respectively. Rxt3p is a subunit of the histone deacetylase Rpd3L, which is recruited to promoters of genes that need to be repressed when galactose is the carbon source. It has been found recently that Rpd3L is necessary for the transcriptional memory of repression i.e. for faster repression rates upon repeated exposure to galactose during carbon source shifts (Lee et al., 2018). Tup1p on the other hand appears to be necessary for long-term transcriptional memory and faster reactivation of galactose inducible GAL genes after a prolonged growth period (>4 h) in glucose that followed initial growth in galactose (Sood et al., 2017). Long-term transcription memory for rapid reactivation of GAL genes requires the Gal1 protein that sequesters the Gal80p repressor in the cytoplasm (Zacharioudakis et al., 2007) and Tup1p-mediated reorganization of the chromatin architecture of GAL gene promoters (Sood et al., 2017). The link between the cytoplasmic activity of Gal1p and the nuclear activity of Tup1p are however unclear, although Tup1p activity appears to be downstream of Gal1p activity.
Although transcriptional memory of earlier exposure to galactose seems to be preserved for several cell generations, the abovementioned studies have not addressed how these factors partition between the mother and the daughter and consequently whether mothers transmit the information to their daughters. Our experiment with cells grown in glucose or galactose (Figure 4) does not directly address whether the retention of “old” Tup1p synthesized before S-phase is involved in transcription memory of galactose metabolism genes and if this transcription memory is confined to the mother cell. It does, however, suggest a possible molecular mechanism for Tup1p-mediated transcriptional memory.
Our results are consistent with a model that restricts Tup1p “tethering” to nuclear structures in the G1 phase, thus ensuring that only Tup1p proteins were present in the cell before replication stay in the mother cell. Interestingly, the localization of GAL genes to the nuclear periphery that occurs during the establishment of GAL transcriptional memory in glucose depends on Tup1p and the Nup100p subunit of the nuclear pore complex (NPC) (Sood et al., 2017). Taken together with the fact that Tup1p binds to the promoters of GAL genes during growth in glucose and galactose (Papamichos-Chronakis et al., 2004; Nehlin et al., 1991), the above observation raises an intriguing possibility that Tup1p may recruit GAL genes to the nuclear periphery via its tether to the NPC. We propose that this hypothetical anchoring of Tup1p induces a conformational change in Tup1p that in turn enhances the interaction of Tup1p with its partners that specifically target Tup1p to gene promoters. Tup1p anchoring is independent of the carbon source, and the putative conformational change caused by anchoring should be generic and should enhance the pairing of Tup1p with any partner, whose presence in the cell on the other hand does depend on the carbon source. Consequently, the task of specifically targeting the tethered Tup1p to repress, activate, or prepare genes for activation in response to glucose, galactose, or during transcription memory establishment falls to the interacting partners. Putative Tup1p tethering is stable throughout the cell cycle and although a fraction of the tethered Tup1p in complex with specific factors will be targeted to genes, another fraction could potentially keep Tup1p partners specific for galactose or glucose or transcription memory in reserve for later use if the carbon source were to shift back and forth. The “new” untethered Tup1p synthesized after G1 would still be functional and would compete with the tethered Tup1p for binding to target genes although possibly with lower efficiency. The untethered Tup1p would also be “free to go” to the daughter cell after mitosis, where it would be tethered to the daughter's nucleus to start a new cycle. If the carbon source were to change during the cell cycle, the tethered and untethered Tup1p that were not bound to partners specific for the first carbon source will be available to bind to partners that respond to the new carbon source, whereas the “reserved” fraction of tethered Tup1p bound to partners responsive to the initial carbon source would be kept as “memory” complexes waiting to be activated if the carbon source were to switch back.
Our analysis of protein decay and synthesis rates has also revealed some unexpected features of protein cycles in budding yeast. It is remarkable that even though protein synthesis and degradation are distinct and seemingly independent processes, they are precisely coordinated. There is actually no protein homeostasis with equal synthesis and decay rates as might be expected. The 2-fold higher synthesis rate ensures instead a steady supply of new proteins. In other words, the level of chromatin proteins increases throughout the cell cycle and never reaches steady state. Consequently, at any given time during the cell cycle newly synthesized “younger” proteins will represent the majority of the total protein population as shown in Figure 5E. Because old proteins are more likely to be damaged, the accumulation of new proteins may be an adaptation to ensure that the cell has optimal amounts of functional proteins at its disposal. Because only about one-third of synthesized proteins whose half-life lasts one cell cycle are degraded in one cell generation, the cell has to rely on dilution through cell division in addition to protein degradation to maintain optimal protein levels. The accumulation of new proteins may also be a hallmark of asymmetrically dividing cells in which the mother cell has to supply all the proteins necessary for her daughter's initial growth until the daughter cell has produced enough proteins on her own.
TrIPP opens up exciting possibilities for investigations of the effects of environmental variability, cell-cycle regulation, and aging processes on the cellular proteome. It will thus be of particular interest to explore the inheritance patterns of transcription factors Adr1p and Xpb1p and redox regulators Srx1p, Sod2p, Tsa2p, and Gpx2p that have all recently been implicated in the regulation of gene expression programs specific to aging cells (Hendrickson et al., 2018). Measurements of a larger protein set are needed to confirm whether the trends we found in our small test set such as the coupling of protein synthesis and degradation that favor the accumulation of new proteins in the cell, half-life durations that match the length of one cell cycle, and the stochastic and symmetric partitioning of proteins between the mother and its daughter represent general trends in protein biology of asymmetrically dividing cells. Likewise, further studies of the mechanisms responsible for the asymmetric repartition of Rxt3p, Tup1p, and Fpr4p proteins should help us better understand the potential role of these proteins in epigenetic inheritance of transcriptional memory.
Limitations of the study
The study used an 18 protein test set as proof of principle of feasibility and as an example of the type of quantitative measurements that can be obtained using TrIPP. Although our results reveal possible tendencies for protein dynamics and inheritance patterns during the cell cycle, we cannot make any generalizations until a larger protein dataset is tested.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Marta Radman-Livaja (marta.radman-livaja@igmm.cnrs.fr).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Data and code availability
All microscopy images and Perl and R scripts are available upon request from the Lead Contact.
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
We thank Virginie Georget and Leslie Bancel-Vallée (MRI, Biocampus, Montpellier) for their invaluable help with the microscope setup. We thank Edouard Bertrand (IGMM, France) for the pDendra2-C plasmid and Angela Taddei (Institut Curie, France) for the pAH52 plasmid. This study has been supported by the ANR 2014 GenChroSeg grant (MRL) (Agence Nationale de la Recherche, France). MA is supported by the Labex EpigenMed fellowship (Laboratoire d'Excellence EpigenMed, University of Montpellier, France).
Author contributions
PV constructed strains and plasmids, optimized and performed the live cell imaging experiments, and analyzed images. MA performed the carbon source shift experiments with Tup1p-dendra2, analyzed images, and did the RT-qPCR and flow cytometry experiments for Tup1p-dendra2 function (Figures 4 and S3). ST and AF constructed strains and prepared samples for mass spectrometry, performed live cell imaging experiments and analyzed images. FRC and IS constructed strains. MRL conceived and designed the experiments, wrote the manuscript, and developed the Perl/R scripts for analysis. KK and SU performed the MS pulse SILAC analysis.
Declaration of interests
The authors declare no competing interests.
Published: February 19, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2021.102075.
Supplemental information
References
- Alber A.B., Paquet E.R., Biserni M., Naef F., Suter D.M. Single live cell monitoring of protein turnover reveals Intercellular variability and cell-cycle dependence of degradation rates. Mol. Cell. 2018;71:1079–1091.e9. doi: 10.1016/j.molcel.2018.07.023. [DOI] [PubMed] [Google Scholar]
- Christiano R., Nagaraj N., Frohlich F., Walther T.C. Global proteome turnover analyses of the Yeasts S. cerevisiae and S. pombe. Cell Rep. 2014;9:1959–1965. doi: 10.1016/j.celrep.2014.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R.T., Struhl G. Causal role for inheritance of H3K27me3 in maintaining the OFF state of a Drosophila HOX gene. Science. 2017;356:eaai8236. doi: 10.1126/science.aai8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoth-Lippuner A., Krzyzanowski M.K., Stober C., Barral Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. Elife. 2014;3:e03790. doi: 10.7554/eLife.03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia Del Arco A., Edgar B.A., Erhardt S. In vivo analysis of centromeric proteins reveals a stem cell-specific asymmetry and an essential role in differentiated, non-proliferating cells. Cell Rep. 2018;22:1982–1993. doi: 10.1016/j.celrep.2018.01.079. [DOI] [PubMed] [Google Scholar]
- Hendrickson D.G., Soifer I., Wranik B.J., Kim G., Robles M., Gibney P.A., McIsaac R.S. A new experimental platform facilitates assessment of the transcriptional and chromatin landscapes of aging yeast. Elife. 2018;7:e39911. doi: 10.7554/eLife.39911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotz M., Leisner C., Chen D., Manatschal C., Wegleiter T., Ouellet J., Lindstrom D., Gottschling D.E., Vogel J., Barral Y. Spindle pole bodies exploit the mitotic exit network in metaphase to drive their age-dependent segregation. Cell. 2012;148:958–972. doi: 10.1016/j.cell.2012.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Keller P.J., Bartosik A., Meurer M., Barry J.D., Mardin B.R., Kaufmann A., Trautmann S., Wachsmuth M., Pereira G. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat. Biotechnol. 2012;30:708–714. doi: 10.1038/nbt.2281. [DOI] [PubMed] [Google Scholar]
- Kornitzer D. Monitoring protein degradation. Methods Enzymol. 2002;351:639–647. doi: 10.1016/s0076-6879(02)51874-7. [DOI] [PubMed] [Google Scholar]
- Laprell F., Finkl K., Muller J. Propagation of Polycomb-repressed chromatin requires sequence-specific recruitment to DNA. Science. 2017;356:85–88. doi: 10.1126/science.aai8266. [DOI] [PubMed] [Google Scholar]
- Lee B.B., Choi A., Kim J.H., Jun Y., Woo H., Ha S.D., Yoon C.Y., Hwang J.T., Steinmetz L., Buratowski S. Rpd3L HDAC links H3K4me3 to transcriptional repression memory. Nucleic Acids Res. 2018;46:8261–8274. doi: 10.1093/nar/gky573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D., Ford T.N., Chu K.K., Metz J. Optically sectioned in vivo imaging with speckle illumination HiLo microscopy. J. Biomed. Opt. 2011;8:016014. doi: 10.1117/1.3528656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader A.W., Richmond R.K., Sargent D.F., Richmond T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- McShane E., Sin C., Zauber H., Wells J.N., Donnelly N., Wang X., Hou J., Chen W., Storchova Z., Marsh J.A. Kinetic analysis of protein stability reveals age-dependent degradation. Cell. 2016;167:803–815 e21. doi: 10.1016/j.cell.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Nehlin J.O., Carlberg M., Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C.J., Santos-Rosa H., Kouzarides T. Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell. 2006;126:905–916. doi: 10.1016/j.cell.2006.07.026. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M., Gligoris T., Tzamarias D. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 2004;5:368–372. doi: 10.1038/sj.embor.7400120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S., Sedkov Y., Johnston D.M., Hodgson J.W., Black K.L., Kovermann S.K., Beck S., Canaani E., Brock H.W., Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M., Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Corrigendum: global quantification of mammalian gene expression control. Nature. 2013;495:126–127. doi: 10.1038/nature11848. [DOI] [PubMed] [Google Scholar]
- Smith R.L., Johnson A.D. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem. Sci. 2000;25:325–330. doi: 10.1016/s0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- Sood V., Cajigas I., D'Urso A., Light W.H., Brickner J.H. Epigenetic transcriptional memory of GAL genes depends on growth in glucose and the Tup1 transcription factor in Saccharomyces cerevisiae. Genetics. 2017;206:1895–1907. doi: 10.1534/genetics.117.201632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagge F., Mitronova G.Y., Belov V.N., Wurm C.A., Jakobs S. SNAP-, CLIP- and Halo-tag labelling of budding yeast cells. PLoS one. 2013;8:e78745. doi: 10.1371/journal.pone.0078745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus K.A., Kaeberlein M., Kennedy B.K. Replicative aging in yeast: the means to the end. Annu. Rev. Cell Dev. Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S., Tanaka M. Distinct segregation patterns of yeast cell-peripheral proteins uncovered by a method for protein segregatome analysis. Proc. Natl. Acad. Sci. U S A. 2019;116:8909–8918. doi: 10.1073/pnas.1819715116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thayer N.H., Leverich C.K., Fitzgibbon M.P., Nelson Z.W., Henderson K.A., Gafken P.R., Hsu J.J., Gottschling D.E. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc. Natl. Acad. Sci. U S A. 2014;111:14019–14026. doi: 10.1073/pnas.1416079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Moazed D. DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science. 2017;356:88–91. doi: 10.1126/science.aaj2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., McCormick M.A., Zheng J., Xie Z., Tsuchiya M., Tsuchiyama S., El-Samad H., Ouyang Q., Kaeberlein M., Kennedy B.K., Li H. Systematic analysis of asymmetric partitioning of yeast proteome between mother and daughter cells reveals "aging factors" and mechanism of lifespan asymmetry. Proc. Natl. Acad. Sci. U S A. 2015;112:11977–11982. doi: 10.1073/pnas.1506054112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharioudakis I., Gligoris T., Tzamarias D. A yeast catabolic enzyme controls transcriptional memory. Curr. Biol. 2007;17:2041–2046. doi: 10.1016/j.cub.2007.10.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All microscopy images and Perl and R scripts are available upon request from the Lead Contact.