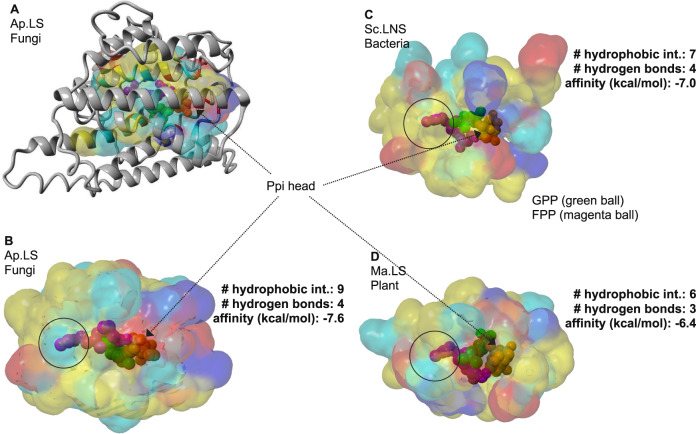
Fig. 6. Structural comparison of linalool synthases in bacteria, fungi, and plants.
A The overlay of 3D structure (gray ribbon) and substrate-binding pocket of Ap.LS. Energy minimized homology model was used with 4LXW as template. GPP and FPP from 2ONG and 6A2C, respectively, were structurally aligned to 4LXW. Binding pocket surfaces for fungal (B), bacterial (C), and plant (D) linalool synthases. GPP (2-fluorogeranyl diphosphate) and FPP ligands are shown as green and magenta spheres, respectively. The solvent-accessible surface of residues in the binding pocket is colored yellow (hydrophobic), cyan (polar), red (negative), or blue (positive). Best docking model from AutoDock Vina was used to submit to the Protein-Ligand Interaction Profiler (PLIP) server (Salentin et al.32). Ap.LS, the most stable protein–ligand complex has the most hydrophobic interactions with the hydrophobic tail of GPP.