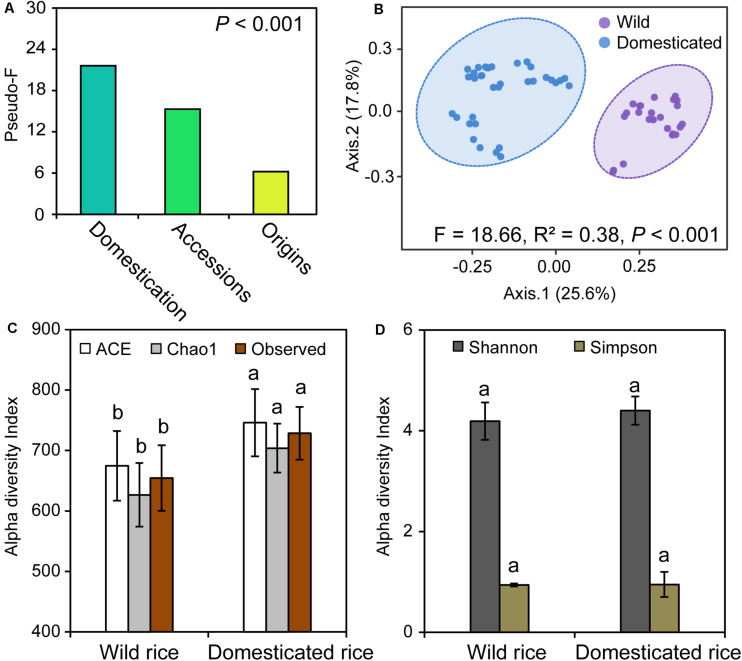
FIGURE 1.
(A) The permutational multivariate analysis of variance (PERMANOVA) output showing the importance of rice domestication, rice species, and origins factors shaping the rhizosphere fungal community. (B) Compositional changes of fungal communities in the rhizospheres of wild and domesticated rice. (C,D) Alpha diversity indices of fungi in the rhizospheres of wild and domesticated rice. The pseudo-F value was used as a proxy for the importance of the factor (A). For the PERMANOVA of domestication factor, all samples were separated into two groups: wild and domesticated rice. For the PERMANOVA of accessions factor, all samples were separated into 12 groups: 12 rice species as described in section “Materials and Methods”. For the PERMANOVA of origins factor, all samples were separated into two groups: African rice and Asian rice. Principal coordinates analysis (PCoA) was performed based on the Bray–Curtis dissimilarity matrix at operational taxonomic units (OTUs) level across all the samples (B). Ellipses demonstrate the mean ± 1 SD, wild rice in purple and domesticated rice in blue. In panels (C,D), wild rice rhizosphere samples, n = 24; domesticated rice rhizosphere samples, n = 35. Different letters within each column indicated significant differences between wild and domesticated rice based on a one-way ANOVA with a Tukey test at the P < 0.05 level.