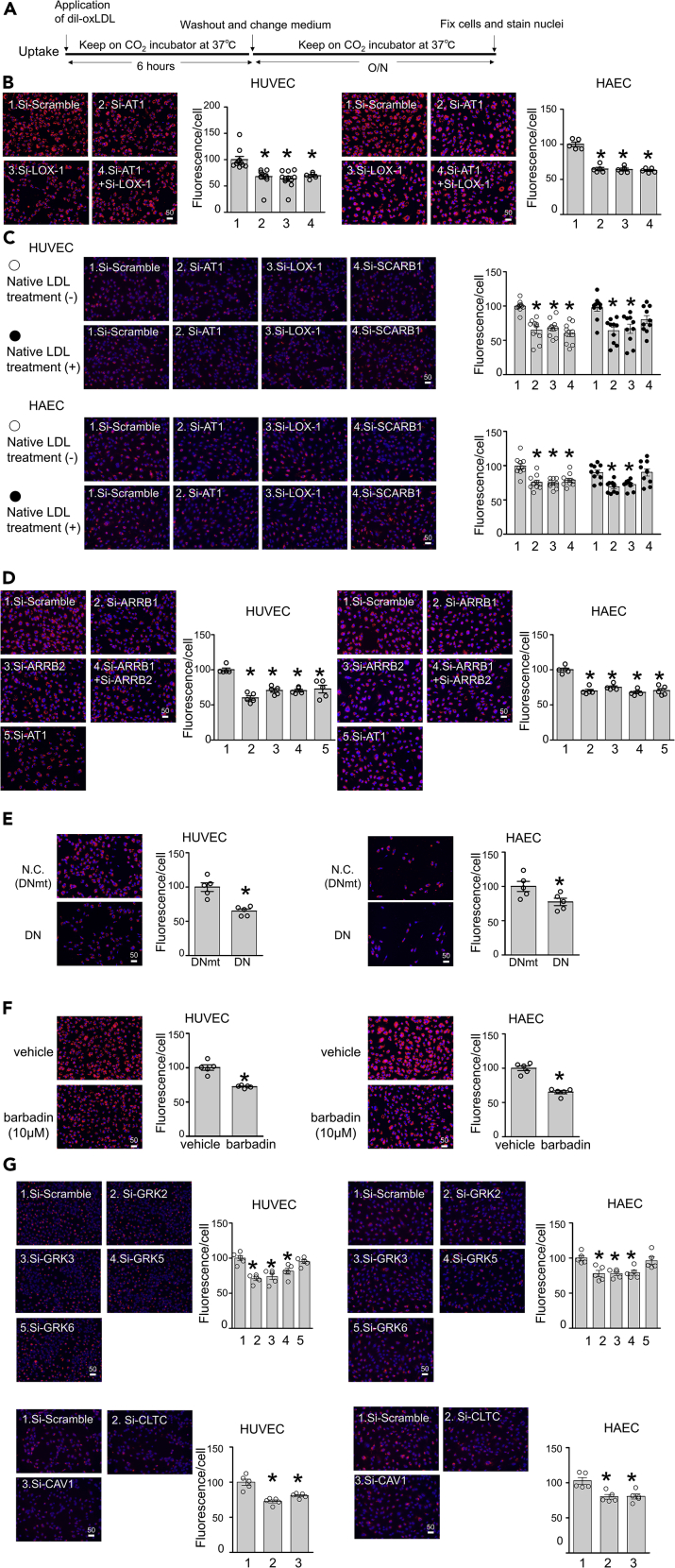
Figure 6.
LOX-1-AT1-β-arrestin-dependent uptake of oxLDL in human endothelial cells
(A) Protocol for visualization of intracellular uptake of Dil-labeled oxLDL in human umbilical vein endothelial cells (HUVECs) and human aortic endothelial cells (HAECs).
(B) Intracellular uptake of 2 μg/mL Dil-labeled oxLDL in cells with siRNA knockdown of indicated genes (n = 5, each). Scale bar, μm. The graph indicates the fluorescence/number of nuclei, and the average value of the group at the far left was normalized to 100%. Data are represented as mean ± SEM. The differences were determined by one-way analysis of variance (ANOVA) with Bonferroni correction. ∗p < 0.01 vs. control. The knockdown efficiency of each gene was demonstrated in Figure S6.
(C) Intracellular uptake of 2 μg/mL Dil-labeled oxLDL in cells with siRNA-mediated knockdown of the indicated genes, pre- and co-treated or not with non-labeled native LDL at 20 μg/mL (n = 10, each). Pretreatment with native LDL was carried out for 1 hr. Scale bar, μm. The graph indicates the fluorescence/number of nuclei, and the average value of the group at the far left was normalized to 100%. Data are represented as mean ± SEM. The differences were determined by one-way ANOVA with Bonferroni correction. ∗p < 0.01 vs. control. The knockdown efficiency of each gene was demonstrated in Figure S6.
(D) Intracellular uptake of 2 μg/mL Dil-labeled oxLDL in cells with siRNA-mediated knockdown of the indicated genes (n = 5, each). Scale bar, μm. The graph indicates the fluorescence/number of nuclei, and the average value of the group at the far left was normalized to 100%.
Data are represented as mean ± SEM. The differences were determined by one-way ANOVA with Bonferroni correction. ∗p < 0.01 vs. control. The knockdown efficiency of each gene was demonstrated in Figure S6.
(E) Intracellular uptake of 2 μg/mL Dil-labeled oxLDL in cells transfected with dominant negative vector of β-arrestin (barre-DN) or negative control vector (N.C.) (n = 5, each). Scale bar, μm. The graph indicates the fluorescence/number of nuclei, and the average value of the group at the far left was normalized to 100%.
Data are represented as mean ± SEM. The differences were determined by Student's t-test. ∗p < 0.01 vs. control (DNmt).
(F) Intracellular uptake of 2 μg/mL Dil-labeled oxLDL in cells pretreated with vehicle or the β-arrestin-specific inhibitor barbadin at 10 μM for 30 min (n = 5, each).
The graph indicates the fluorescence/number of nuclei, and the average value of the group at the far left was normalized to 100%. Scale bar, μm.
The graph indicates the fluorescence/number of nuclei, and the average value of the group at the far left was normalized to 100%.
Data are represented as mean ± SEM. The differences were determined by Student's t-test. ∗p < 0.01 vs. control.
(G and H) Intracellular uptake of 2 μg/mL Dil-labeled oxLDL in cells with siRNA-mediated knockdown of the indicated genes (n = 5, each). Scale bar, μm.
The graph indicates the fluorescence/number of nuclei, and the average value of the group at the far left was normalized to 100%. Data are represented as mean ± SEM. The differences were determined by one-way ANOVA with Bonferroni correction. ∗p < 0.01 vs. control. The knockdown efficiency of each gene was demonstrated in Figure S6.