The CckA, ChpT, and CtrA phosphorelay proteins are widespread in the alphaproteobacteria, and there are two groups of organisms that differ in terms of whether this pathway is essential for cell viability. Little is known about the biochemical function of these proteins in organisms where the pathway is not essential, a group that includes Rhodobacter capsulatus.
KEYWORDS: Rhodobacter, CckA, ChpT, CtrA, cyclic di-GMP, phosphorelay, Phos-tag gel, gene transfer agent, RcGTA
ABSTRACT
Protein phosphorylation is a universal mechanism for transducing cellular signals in prokaryotes and eukaryotes. The histidine kinase CckA, the histidine phosphotransferase ChpT, and the response regulator CtrA are conserved throughout the alphaproteobacteria. In Rhodobacter capsulatus, these proteins are key regulators of the gene transfer agent (RcGTA), which is present in several alphaproteobacteria. Using purified recombinant R. capsulatus proteins, we show in vitro autophosphorylation of CckA protein, and phosphotransfer to ChpT and thence to CtrA, to demonstrate biochemically that they form a phosphorelay. The secondary messenger cyclic di-GMP changed CckA from a kinase to a phosphatase, resulting in reversal of the phosphotransfer flow in the relay. The substitutions of two residues in CckA greatly affected the kinase or phosphatase activity of the protein in vitro, and production of mutant CckA proteins in vivo confirmed the importance of kinase but not phosphatase activity for the lytic release of RcGTA. However, phosphatase activity was needed to produce functional RcGTA particles. The binding of cyclic di-GMP to the wild-type and mutant CckA proteins was evaluated directly using a pulldown assay based on biotinylated cyclic di-GMP and streptavidin-linked beads.
IMPORTANCE The CckA, ChpT, and CtrA phosphorelay proteins are widespread in the alphaproteobacteria, and there are two groups of organisms that differ in terms of whether this pathway is essential for cell viability. Little is known about the biochemical function of these proteins in organisms where the pathway is not essential, a group that includes Rhodobacter capsulatus. This work demonstrates biochemically that CckA, ChpT, and CtrA also form a functional phosphorelay in the latter group and that the direction of phosphotransfer is reversed by cyclic di-GMP. It is important to improve understanding of more representatives of this pathway in order to obtain deeper insight into the function, composition, and evolutionary significance of a wider range of bacterial regulatory networks.
INTRODUCTION
All cells sense and respond to environmental and intracellular changes, and a common mechanism for such responses in bacteria is histidyl-aspartyl phosphorelay systems composed of two or more proteins (1). Such systems operate as pathways that typically begin with a sensor protein that autophosphorylates on a histidine residue and end with a response regulator protein that carries out an activity. Many response regulators bind DNA differentially depending on whether they are phosphorylated on an aspartate residue. The phosphorelay pathway from the histidine kinase CckA (2) through the histidine phosphotransferase ChpT (3) to the response regulator CtrA (4) was discovered in Caulobacter crescentus, where it functions in DNA replication and cell cycle progression and is essential for viability. Homologues of these proteins are widespread in the alphaproteobacteria, but few of these putative pathways have been studied in great detail. Although the C. crescentus system stands as the premier model for a variety of organisms, it is clear that in addition to similarities, there are differences in the roles and functioning of this pathway among different species, perhaps due to differences related to symmetrical versus asymmetrical cell division processes (5–10).
The signal(s) sensed by a CckA protein had been elusive until it was shown that the DivL protein controls C. crescentus CckA kinase/phosphatase activity differentially depending on the phosphorylation state of DivK (11) and, subsequently, that the second messenger cyclic diguanylate (c-di-GMP) binds directly to the CckA of C. crescentus and Agrobacterium tumefaciens (12). In the latter work, Lori et al. also showed that a consequence of CckA binding of c-di-GMP in C. crescentus and A. tumefaciens is to reverse the phosphorelay, with CckA acting as a phosphatase instead of a kinase. The potential for dephosphorylation of a response regulator because of a change in the activity of a histidine kinase had been known for years (13, 14), and c-di-GMP was the first of several cyclic dinucleotides found to control cellular processes (15, 16). In C. crescentus specifically, although not essential for the cell cycle to function, c-di-GMP acts as a major driver of cell division and morphogenesis (17, 18).
We discovered the Rhodobacter capsulatus ctrA (rcc01663) and cckA (rcc01749) genes in screens to identify mutations that affect the production of the R. capsulatus gene transfer agent (RcGTA) (19), a prophage-like entity encoded by genes scattered over the chromosome, which transfers random segments of the genome and is released by cell lysis (20). Subsequently, a gene encoding a weak homologue of ChpT (rcc03000) was identified, and it was found that a deletion of the chpT gene, as well as mutations of the cckA and ctrA genes, affected the production of RcGTA particles and their release from cells (21). This signaling system also regulates R. capsulatus flagellar motility at the level of flagellar gene expression (8, 21, 22). The sequence identity between the C. crescentus and R. capsulatus CtrA homologues is high, at 71%. The N-terminal regions of the CckA proteins do not align well, but the regions of the proteins associated with phosphotransfer reactions are 44% identical. Similarly, the ChpT proteins are only 28% identical over their full lengths but have a small region centered around the histidine involved in phosphotransfer where they are 68% identical (19, 21). However, autophosphorylation of the R. capsulatus CckA protein and the transfer of phosphate to ChpT and onward to CtrA have not been demonstrated biochemically.
In total, the R. capsulatus genome contains 20 genes encoding proteins with domains associated with the synthesis (diguanylate cyclases) or degradation (phosphodiesterases) of c-di-GMP (23). Eight of these genes were identified as having lower transcript levels in a ctrA-null mutant strain, suggesting possible interactions between the CtrA phosphorelay system and signaling via c-di-GMP (8). Indeed, it was recently shown that four of these genes affect RcGTA gene expression and production as well as flagellar motility: evaluation of the activities of the c-di-GMP-related domains in the four proteins, expression of heterologous diguanylate cyclase- and phosphodiesterase-encoding genes, and quantification of intracellular c-di-GMP levels demonstrated that c-di-GMP inhibits RcGTA production in R. capsulatus (24). However, a mechanistic connection of c-di-GMP levels to RcGTA was not demonstrated.
Although the putative CckA-ChpT-CtrA pathway of R. capsulatus shares several features (e.g., control of flagellar motility) with the C. crescentus system, a major difference is that the genes encoding these proteins in R. capsulatus are not essential for cell viability. Such differences from the C. crescentus paradigm in many species have been noted and may stem from the lack of some components (such as the DivJ-PleC-DivK and RcdA-CpdR systems) of the broad regulatory network that exists in C. crescentus (5, 8, 25, 26). Because of such differences, it is important to improve the understanding of a variety of CckA, ChpT, and CtrA proteins in order to progress toward a deeper understanding of the function, composition, and evolutionary significance of a wider range of bacterial regulatory networks.
Here, we describe the use of purified R. capsulatus proteins to demonstrate in vitro autophosphorylation of CckA and the transfer of phosphate to ChpT and thence to CtrA, yielding phosphorylated CtrA (CtrA∼P). We went on to use this in vitro assay to show ChpT- and CckA-dependent dephosphorylation of CtrA∼P and to investigate whether c-di-GMP and ADP affect the direction of phosphate transfer through this pathway. Key amino acids in CckA, chosen on the basis of conservation with residues that affect the kinase or phosphatase activity of the C. crescentus protein, were mutated in order to investigate the effects of these mutations on R. capsulatus CckA activities in vitro and on RcGTA-related cell lysis when expressed in vivo.
RESULTS
In vitro phosphorylation of R. capsulatus CckA, ChpT, and CtrA.
We expressed recombinant R. capsulatus ctrA and chpT genes that encode 6 His residues added to the N terminus, as well as a cckA construct that encodes a soluble version of CckA lacking the N-terminal 69 amino acids, which include the two transmembrane segments, and containing a 6His tag added to the C terminus. The purified proteins were incubated singly or in various combinations in the presence or absence of ATP or acetyl phosphate (acetyl∼P) in order to determine whether the proteins are capable of autophosphorylation and transfer of phosphate to another protein. The phosphorylation state of proteins was determined by their mobility in a Phos-tag gel, where the electrophoretic mobility of a protein is decreased as a result of phosphorylation (27). Although all of the proteins studied in this in vitro work were recombinant 6His-tagged proteins, we refer to them here simply as CckAΔ69, ChpT, and CtrA.
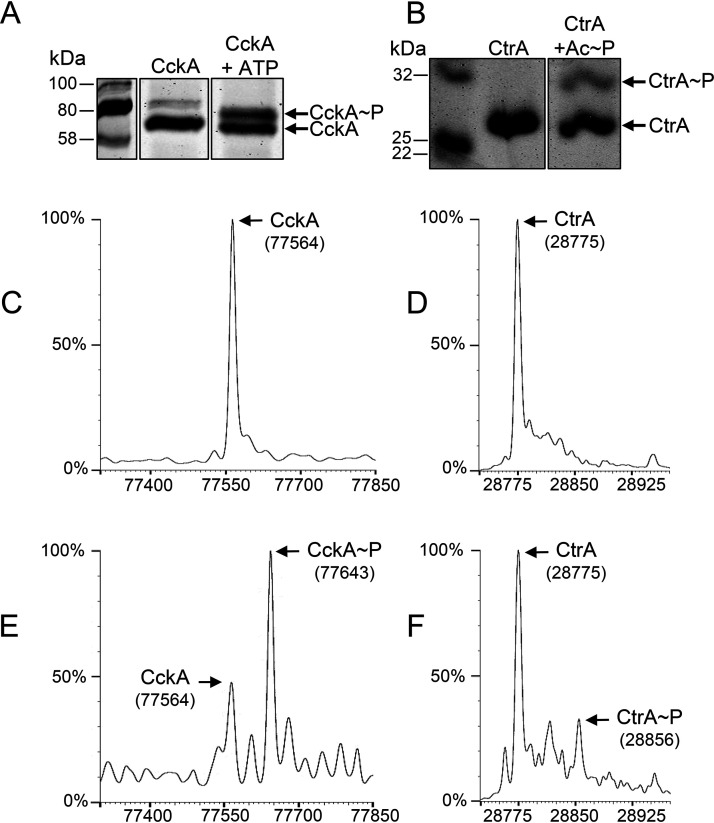
CckAΔ69 was capable of autophosphorylation using ATP as the phosphate donor, as indicated by the change in the banding pattern in a Phos-tag gel (Fig. 1A; see also Fig. S1 in the supplemental material), and the phosphorylation was confirmed by the appearance of a peak in the mass spectrum that differed by the ∼80-Da mass of a phosphate from the 77,564-Da peak attributed to CckAΔ69 (compare Fig. 1C and E).
FIG 1.
R. capsulatus CckA and CtrA are capable of autophosphorylation, and the phosphorylated proteins are separated from nonphosphorylated proteins in the Phos-tag gel system. (A) Gel migration of CckA versus CckA∼P. (B) Gel migration of CtrA versus CtrA∼P. (C) Mass spectrum of a CckA sample prior to phosphorylation with ATP. (D) Mass spectrum of a CtrA sample prior to phosphorylation with acetyl∼P. (E) Increase in the mass of CckA after incubation with ATP. (F) Increase in the mass of CtrA after incubation with acetyl phosphate.
The ability of CtrA to be phosphorylated was initially assessed by using acetyl∼P as a phosphate donor, as has been used for the autophosphorylation of similar response regulators (28). Figure 1B and Fig. S2 show that a slower-migrating band appeared on a Phos-tag gel after the incubation of CtrA with acetyl∼P, indicating that this protein autophosphorylated in the presence of this phosphate donor. Although the phosphorylation was partial, the appearance of a new protein with a mass 80 Da greater than that of the CtrA peak was confirmed by mass spectrometry (compare Fig. 1D and F). This information was used in other experiments described below.
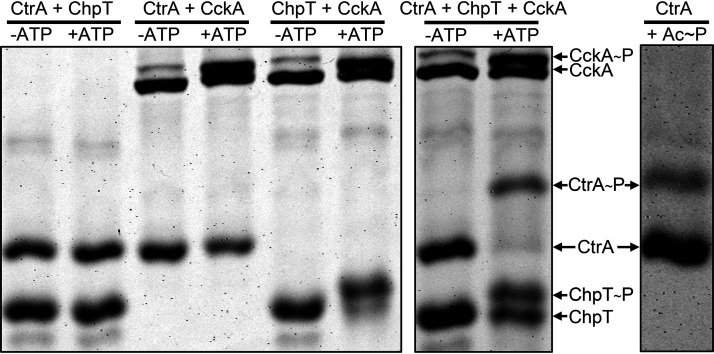
Figure 2 shows the Phos-tag gel mobilities of the CckAΔ69, ChpT, and CtrA proteins incubated pairwise in the absence or presence of ATP. Again, CckAΔ69 exhibited an ATP-dependent Phos-tag gel mobility shift, whereas neither ChpT nor CtrA was capable of ATP-dependent autophosphorylation in the absence of CckAΔ69. When CckAΔ69 and CtrA were incubated together in the presence of ATP, CckAΔ69 appeared to autophosphorylate, but there was no change in the gel mobility of CtrA. This result shows that CckAΔ69∼P is incapable of direct transfer of phosphate to CtrA.
FIG 2.
ATP-dependent phosphate transfer in vitro from CckA∼P to CtrA requires ChpT. Proteins were incubated pairwise or all together in the presence or absence of ATP, as indicated above the lanes of the Phos-tag gel. For comparison, a Phos-tag gel of CtrA∼P produced by incubation with acetyl∼P is shown on the right.
However, when CckAΔ69 and ChpT were incubated together in the presence of ATP, there was a clear decrease in the mobility of ChpT, in addition to the previously noted change in the CckA banding pattern (Fig. 2). These results indicate that phosphorylated CckA (CckA∼P) transfers phosphate directly to ChpT, in contrast to CtrA.
When CckAΔ69, ChpT, and CtrA were incubated together in the presence of ATP, it was found that, in addition to CckAΔ69 and ChpT phosphorlyation, CtrA exhibited a gel mobility shift due to phosphorylation (Fig. 2). Taken together, these experiments show that these CckAΔ69, ChpT, and CtrA proteins form a phosphorelay in vitro and are therefore likely to engage in the transfer of phosphate from ATP to CtrA mediated by CckA and ChpT in vivo.
As a control, samples treated as in the experiment for which results are shown in Fig. 2 were run in a standard SDS-PAGE gel (Fig. S3), in which a gel mobility change would require a ≥2-kDa change in protein mass to be detectable. Neither CckAΔ69, ChpT, nor CtrA exhibited a change in mobility in standard SDS-PAGE gels, a result consistent with the interpretation that the mobility shift seen in the Phos-tag gels was due not to a major change in mass or conformation but to the addition of a phosphate moiety to the proteins, resulting in a decrease in gel mobility specifically because of interaction with the phosphate chelator present in the Phos-tag gel.
The data presented above show that the phosphorylation status of CtrA when incubated in the presence of CckAΔ69 and ChpT can be used to reveal the enzymatic activity of CckA. In the following sections, we use the phosphorylation status of CtrA when incubated in the presence of CckAΔ69 and ChpT as an assay of the relative kinase and phosphatase activities of CckA. This assay was applied to wild-type (WT) CckAΔ69 in the presence of the potential effectors c-di-GMP and ADP, as well as to mutant CckAΔ69 proteins containing specific amino acid substitutions.
The CckA protein acts as a phosphatase in the presence of c-di-GMP to reverse the direction of the phosphorelay.
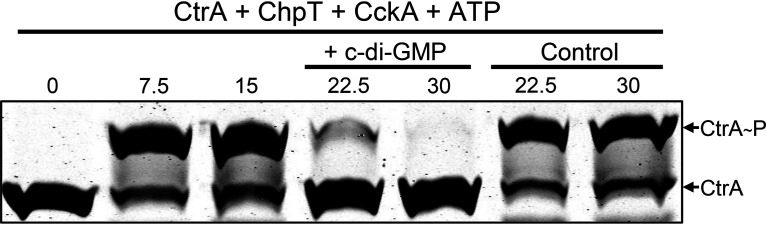
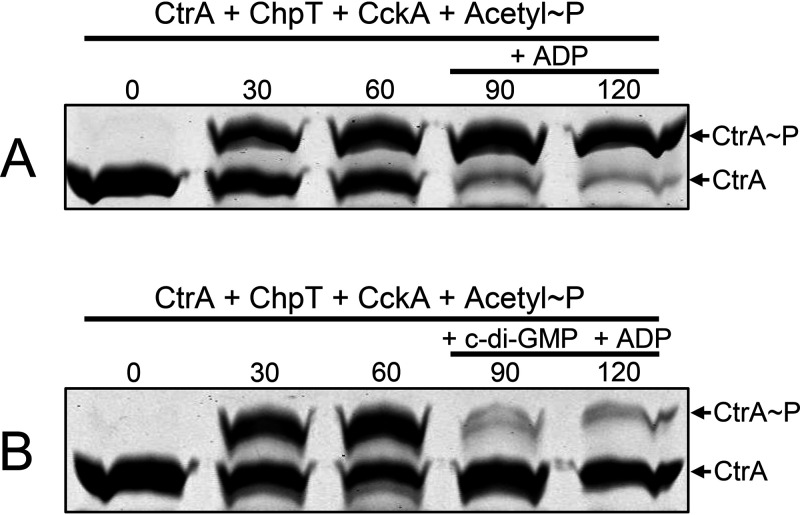
The production of RcGTA is affected by mutations in diguanylate cyclase- and phosphodiesterase-encoding genes, and a variety of histidine kinases, including C. crescentus CckA, have been shown to bind c-di-GMP and change from a kinase to a phosphatase (29, 30). Therefore, we investigated whether R. capsulatus CckA might respond to the presence of c-di-GMP in a similar way. We initially used a kinetic approach in which CtrA was phosphorylated when incubated with the CckAΔ69 and ChpT proteins and ATP. After sufficient time had elapsed to allow for the formation of a substantial amount of CtrA∼P, half of the reaction mixture was transferred to a second tube containing c-di-GMP (5 µM), and samples were removed from both tubes at time intervals to determine the relative amounts of CtrA∼P and CtrA.
As shown in Fig. 3, the phosphorelay rapidly converted most of the CtrA to CtrA∼P within 7.5 min. The reaction mixture was divided in half at 15 min; c-di-GMP was added to one of the two reaction mixtures, and additional samples were removed for Phos-tag gel analysis at the 22.5- and 30-min time points. The amount of CtrA∼P decreased in the presence of c-di-GMP (Fig. 3; Fig. S4), whereas in the absence of c-di-GMP, the amount of CtrA∼P changed little over the same time span. These results show that c-di-GMP reverses the direction of the R. capsulatus CckA-ChpT-CtrA pathway from phosphorylation of CtrA to dephosphorylation of CtrA∼P. There was a decrease in the CckAΔ69∼P Phos-tag gel band accompanying the decrease in the CtrA∼P band after the addition of c-di-GMP (Fig. S5), so it appears that the CckA phosphatase activity rapidly converts CckA∼P to CckA and inorganic phosphate.
FIG 3.
Effect of the addition of c-di-GMP on CtrA phosphorylation in a phosphorelay assay. The presence of c-di-GMP causes a change from phosphorylation of CtrA to dephosphorylation of CtrA∼P. Samples were removed at the indicated time intervals (given in minutes); after 15 min, the reaction mixture was split, and c-di-GMP was added to one of two tubes.
ADP does not stimulate R. capsulatus CckA phosphatase activity.
A variety of histidine kinases change their relative kinase and phosphatase activities in response to ADP (31), and the binding of ADP in concert with c-di-GMP stimulates the phosphatase activity of C. crescentus CckA, apparently because the binding of ADP stabilizes the binding of c-di-GMP (17, 32). In the experiments represented by Fig. 3, ADP was produced by the kinase activity of CckAΔ69, so it was possible that a combination of ADP and c-di-GMP is needed for conversion of R. capsulatus CckA from a kinase to a phosphatase. We used several approaches to investigate the possibility of an effect of ADP on the phosphatase activity of CckAΔ69 and to determine whether ADP affects the binding of c-di-GMP.
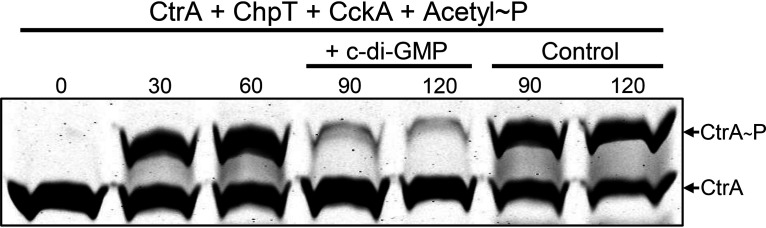
In one type of experiment, CckAΔ69, ChpT, and CtrA were incubated together with acetyl∼P in place of ATP to allow for the formation of CtrA∼P, after which one-half of the reaction mixture was transferred to a second tube containing c-di-GMP, and samples were removed from both tubes at time intervals to determine the relative amounts of CtrA∼P and CtrA. The use of acetyl∼P does not result in the production of ADP, and therefore, CckAΔ69 incubated in the presence of CtrA∼P obtained by acetyl∼P-phosphorylation should lack ADP. As shown in Fig. 4, the addition of c-di-GMP to a mixture of the phosphorelay components in the absence of ATP (and hence ADP) resulted in the dephosphorylation of CtrA∼P, whereas the control reaction mixtures lacking c-di-GMP did not exhibit significant dephosphorylation. The time scale of this experiment is longer than that of the experiment for which results are shown in Fig. 3 because of the competition between dephosphorylation by CckAΔ69 and phosphorylation by acetyl∼P. These results show that the phosphorelay reverses direction in the presence of c-di-GMP and in the absence of ADP and that phosphorylation by acetyl∼P is slower than dephosphorylation driven by CckAΔ69.
FIG 4.
The addition of c-di-GMP to the phosphorelay components in the absence of ATP results in the dephosphorylation of CtrA∼P. CtrA was phosphorylated with acetyl∼P. Samples were removed at the indicated time intervals (given in minutes); after 60 min, the reaction mixture was split, and c-di-GMP was added to one of the two tubes.
Other experiments on CtrA∼P obtained by phosphorylation with acetyl∼P showed that c-di-GMP did not stimulate dephosphorylation of CtrA∼P in the absence of either CckAΔ69 or ChpT (Fig. S6). Therefore, the dephosphorylation of CtrA∼P involves a genuine reversal of the phosphorelay, with ChpT needed to transfer phosphate from CtrA∼P to CckA. Because there was no significant dephosphorylation of CtrA∼P when incubated with ChpT alone, it appears that the driving force results from CckA activity: a high ratio of ChpT to ChpT∼P resulting from CckA phosphatase activity drives the dephosphorylation of CtrA∼P, whereas a high ratio of ChpT∼P to ChpT resulting from CckA kinase activity drives the phosphorylation of CtrA.
In other experiments to address the possible effect of ADP on the direction of the phosphorelay, ADP was added to reaction mixtures containing CckAΔ69, ChpT, and CtrA, in which CtrA∼P was obtained using acetyl∼P. As shown in Fig. 5A, the addition of ADP in the absence of c-di-GMP did not result in detectable phosphatase activity. Furthermore, the addition of ADP and c-di-GMP (Fig. 5B) did not result in discernible stimulation of the phosphatase activity relative to the addition of c-di-GMP alone (compare Fig. 4 and Fig. 5B).
FIG 5.
CckA phosphatase activity is not activated by ADP alone, nor is c-di-GMP-activated phosphatase affected by the presence of ADP. CtrA was phosphorylated with acetyl∼P. Samples were removed at the times indicated (in minutes), with the addition of ADP alone (A) or ADP plus c-di-GMP (B) after 60 min.
Although it is possible that ADP has a small effect on the R. capsulatus CckA kinase versus phosphatase activity, such an effect, if it exists, is dwarfed by the effect of c-di-GMP. To further investigate a possible effect of ADP, we turned to a direct measurement of the binding of c-di-GMP to CckAΔ69. This approach utilized an excess of biotin-tagged c-di-GMP that was mixed with CckAΔ69, and the amount of CckAΔ69 bound to the tagged c-di-GMP was measured by pulldown using streptavidin-linked agarose beads, followed by Western blotting of pulled-down protein using an anti-6His antiserum to detect the His-tagged CckAΔ69 protein.
Compared to the amount of CckAΔ69 that was pulled down in the absence of biotinylated c-di-GMP, there was an 8.6-fold increase after the addition of biotinylated c-di-GMP (Fig. 6); therefore, CckAΔ69 readily binds c-di-GMP in the absence of ADP. However, there was a 1.6-fold increase in the amount of CckAΔ69 pulled down in the presence of c-di-GMP and ADP relative to the amount pulled down in the presence of c-di-GMP alone, so it appears that ADP has a small effect on the binding of c-di-GMP.
FIG 6.
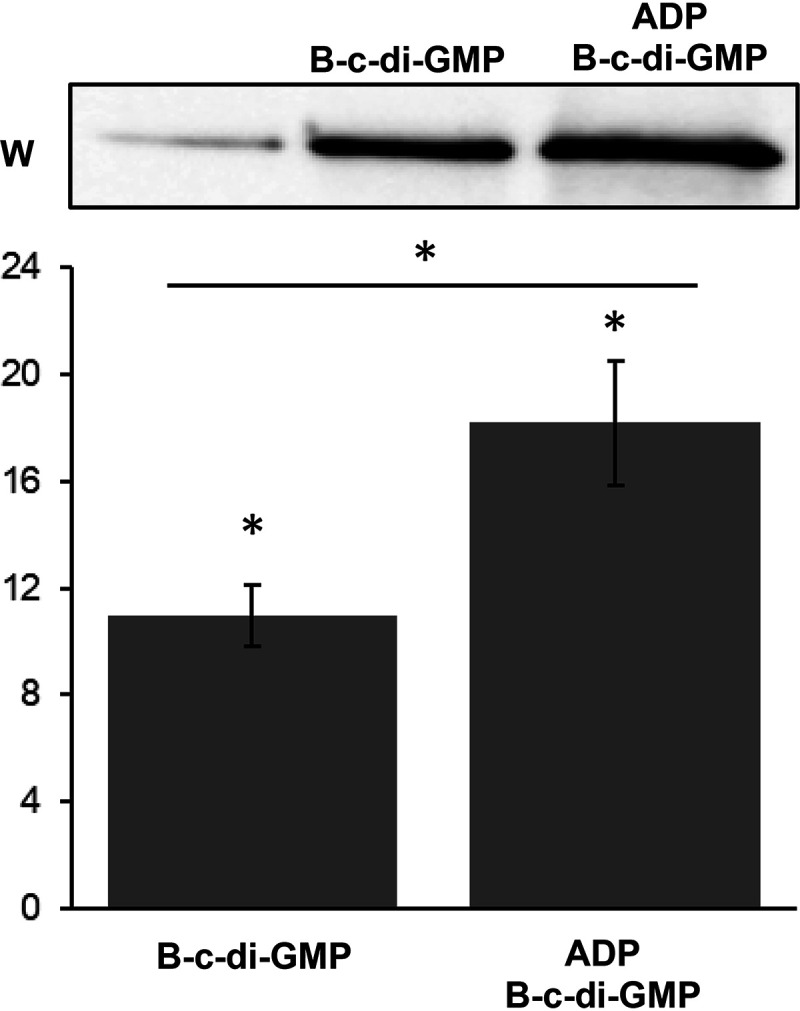
Pulldown of CckA using biotin-tagged c-di-GMP (B-c-di-GMP) shows that ADP weakly stimulates the binding of c-di-GMP. The vertical axis of the graph gives the fold change in band intensity relative to the band intensity of the negative control (beads and CckA alone), shown in the leftmost lane of the gel. Error bars indicate standard deviations (n = 6). Statistical differences (P < 0.05) indicated by asterisks were identified by by one-way ANOVA followed by Tukey’s honestly significant difference post hoc analysis.
These pulldown results confirm the Phos-tag gel mobility data. Together, they indicate that c-di-GMP is bound by CckA and converts the enzymatic activity from that of a kinase to that of a phosphatase, whereas ADP only weakly stimulates the binding of c-di-GMP, a finding consistent with the absence of a significant effect of ADP on the relative kinase versus phosphatase activity of CckA.
CckA kinase and phosphatase activities can be separated by substitutions of conserved residues H399 and Y589, and Y589 is required for the binding of c-di-GMP.
Amino acid residues required for the kinase and phosphatase activities of CckA have been identified in the C. crescentus protein and homologues, some of which have been shown to be involved in the binding of c-di-GMP and ADP (17, 29, 32–34). Although there is a high degree of sequence identity of such residues and the flanking sequences in the R. capsulatus CckA (Fig. S7), it was possible that these residues do not contribute to CckA activities as they do in C. crescentus. Therefore, to address this question, we created two CckAΔ69 proteins containing mutations that affected kinase and phosphatase activities in the C. crescentus homologue. The mutants we created, using R. capsulatus numbering and predicted phenotypes based on the C. crescentus mutant proteins, are the H399A (predicted to lack kinase activity and to retain phosphatase activity) and Y589D (predicted to retain kinase activity and to lack phosphatase activity) proteins. These mutant proteins were evaluated for ATP-dependent kinase activity and phosphotransfer to ChpT and onward to CtrA, as well as for phosphatase activity after the addition of c-di-GMP to the same reaction mixture. The phosphatase activity was also evaluated by phosphorylating CtrA with acetyl∼P, followed by the addition of c-di-GMP and ADP.
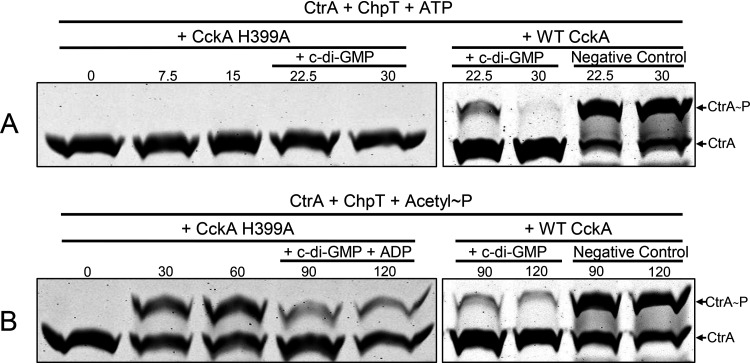
The CckAΔ69 H399A mutant protein lacks kinase activity, as evidenced by the fact that the Phos-tag gel banding pattern of CtrA did not change during 15 min of incubation in the presence of the mutant CckAΔ69, ChpT, and ATP (Fig. 7A). There was no change in the Phos-tag gel banding pattern of the CckAΔ69 H399A mutant protein when incubated in the presence of ATP (Fig. S8), and the addition of c-di-GMP did not affect the migration of CckAΔ69 or CtrA. Because the CckAΔ69 H399A protein lacks kinase activity and therefore does not transfer phosphate from ATP to ChpT and onward to CtrA, it was not possible to use this approach to determine whether the addition of c-di-GMP resulted in reversal of the ATP-driven phosphotransfer pathway from CckA to CtrA. Therefore, we used acetyl∼P to phosphorylate CtrA for 60 min, followed by the addition of c-di-GMP (and ADP, in the presence of both CckAΔ69 and ChpT) to the reaction mixture (Fig. 7B). There was a decrease in the CtrA∼P band intensity after the addition of c-di-GMP, so the CckAΔ69 H399A mutant protein retained c-di-GMP-stimulated phosphatase activity (compare Fig. 7B with the data obtained with WT CckAΔ69 in Fig. 5B).
FIG 7.
The CckA H399A mutant protein lacks kinase activity but retains c-di-GMP-stimulated phosphatase activity. Samples were removed at the times indicated (in minutes), with the addition of c-di-GMP alone after 15 min (A) or ADP plus c-di-GMP after 60 min (B). WT CckA controls from the experiments presented in Fig. 3 and 4 are shown on the right for comparison.
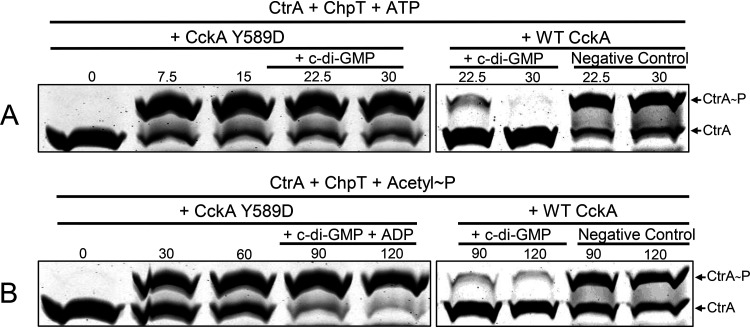
The CckAΔ69 Y589D mutant protein retained kinase activity, as evidenced by the fact that the banding patterns of CckAΔ69, ChpT, and CtrA all changed as in WT CckAΔ69 in response to the presence of ATP (Fig. 8A). However, after the addition of c-di-GMP to reaction mixtures, there was no decrease in the intensity of the CtrA∼P band, regardless of whether CtrA∼P was formed by the phosphorelay with ATP as the phosphate donor or with acetyl∼P as the donor (Fig. 8). Therefore, the Y589D mutant lacks phosphatase activity, as do the CckA proteins of C. crescentus and A. tumefaciens (12).
FIG 8.
The Y589D mutant CckA protein retains kinase activity but lacks c-di-GMP-stimulated phosphatase activity. Samples were removed at the times indicated (in minutes), with the addition of c-di-GMP alone after 15 min (A) or ADP plus c-di-GMP after 60 min (B). WT CckA controls from the experiments presented in Fig. 3 and 4 are shown on the right for comparison.
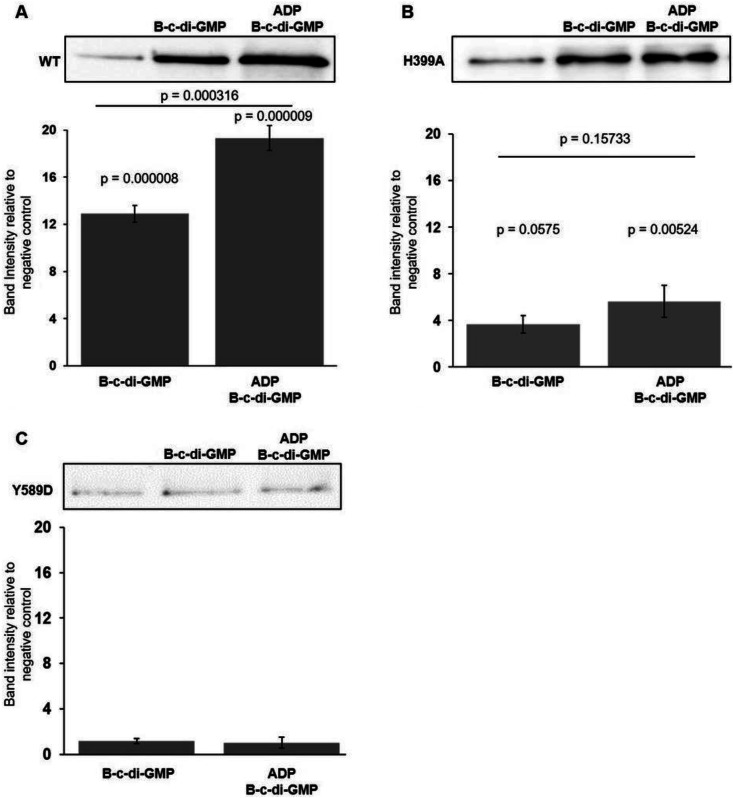
Because the Phos-tag gel migration data did not show an obvious effect of the addition of ADP to CckA kinase and phosphatase assay mixtures containing WT CckAΔ69 (Fig. 5), whereas it was found that the presence of ADP stimulated the binding of c-di-GMP 1.6-fold (Fig. 6), it was of interest to use this direct measurement of the binding of c-di-GMP and the effect of ADP to better understand the effects of site-directed mutations on CckA activities. Biotinylated c-di-GMP was mixed with CckAΔ69 mutant proteins, and streptavidin bead pulldown followed by Western blotting was used to assess the binding of c-di-GMP to CckAΔ69 in the presence and absence of ADP.
As shown in Fig. 9A, WT CckAΔ69 protein bound biotinylated c-di-GMP, resulting in a 13-fold increase in the amount of CckAΔ69 pulled down by streptavidin beads over the amount pulled down in the absence of c-di-GMP. The amount of the H399A protein pulled down in the presence of biotinylated c-di-GMP was less than a 4-fold increase over the amount pulled down in the absence of biotinylated c-di-GMP (Fig. 9B), or less than 30% of the relative amount pulled down using WT CckAΔ69. The Y589D CckAΔ69 mutant did not bind c-di-GMP, as evidenced by the fact that the amount of CckAΔ69 pulled down did not increase in the presence of c-di-GMP (Fig. 9C).
FIG 9.
Pulldown assay of the binding of c-di-GMP to mutant CckA proteins and the effect of ADP. Western blots of His-tagged CckA proteins pulled down using streptavidin beads probed with 6His antisera are shown for each of three strains, above graphs showing the average pixel densities of Western blot bands normalized to the band intensity of the negative control (in the absence of biotinylated c-di-GMP [B-c-di-GMP]). (A) WT CckA protein; (B) the CckA H399A protein; (C) the CckA Y589D protein. Error bars indicate standard deviations (n = 3). Statistical analysis was performed by one-way ANOVA, followed by Tukey’s honestly significant difference post hoc analysis.
The presence of ADP in the pulldown experiments on WT CckAΔ69 increased the amount of CckAΔ69 pulled down about 1.4-fold. The effect of ADP on the pulldown of mutant CckA proteins was not statistically significant, although the average of the pulldown values for the H399A protein was 1.5-fold greater in the presence of ADP.
The data obtained using the Y589D mutant are consistent with the inhibitory effect on phosphatase activity and c-di-GMP binding reported for a homologous C. crescentus mutant CckA protein (17, 29). However, because the H399A mutation did not greatly affect phosphatase activity (Fig. 7), the decrease in the H399A pulldown band to 37% of the WT value (Fig. 9) shows that this degree of inhibition of c-di-GMP binding has a minor effect on the switch of R. capsulatus CckA from kinase to phosphatase activity.
CckA kinase activity, but not phosphatase activity, is important for cell lysis needed for the release of RcGTA.
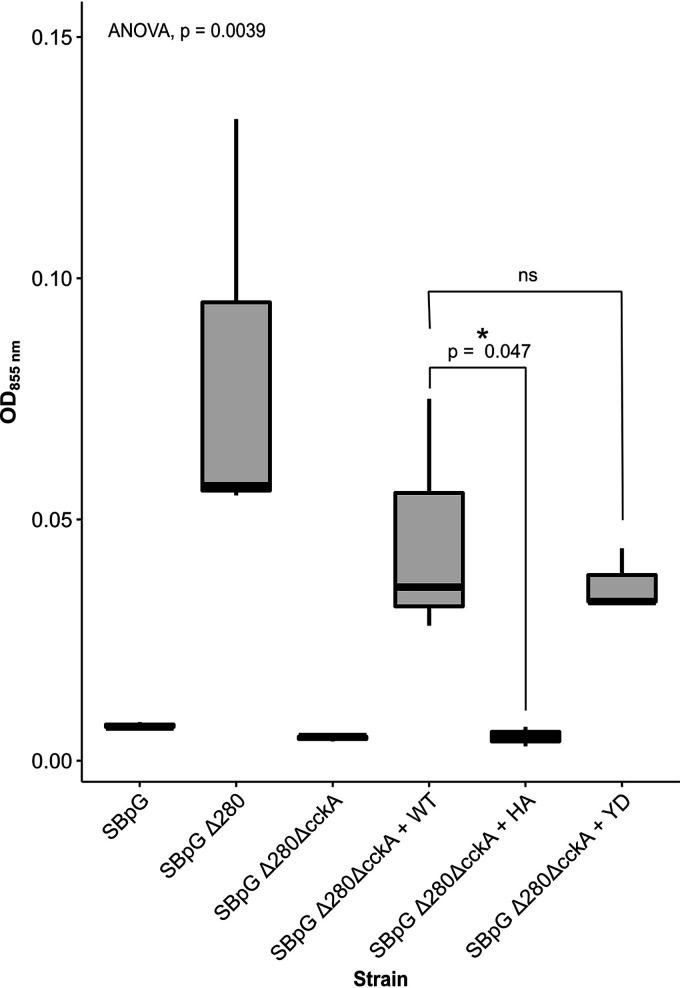
We showed previously that the absence of CckA abolishes cell lysis associated with the release of RcGTA (21, 35), but the relative effects of CckA kinase versus phosphatase activity on cell lysis were unknown. To address this question, the residue changes in the mutant CckAΔ69 proteins (H399A and Y589D) that we describe here in the in vitro kinase/phosphatase assays were created by mutagenesis of the cckA gene encoding the full-length protein, and plasmid-borne copies of these genes were expressed in a strain in which 91% of the cckA gene had been deleted. Cultures were grown to the stationary phase to induce cell lysis accompanying the production of RcGTA, and the degree of cell lysis was measured by the release of photosynthetic pigments into the cell-free culture medium. To facilitate the detection of pigments, the RcGTA overproducer strain SBpGΔ280 was used as the background (36). As shown in Fig. 10, the overproducer strain SBpGΔ280 released a much greater amount of normally intracellular pigments (measured by absorption at 855 nm) than the parental strain SBpG.
FIG 10.
Effects of CckA residue changes H399A (HA) and Y589D (YD) on cell lysis. Increased cell lysis results in increased optical density at 855 nm (OD855). The results of one-way ANOVA [F(5,12) = 6.46, P = 0.004] and t tests of two a priori hypotheses are indicated (n = 3). (The asterisk indicates significance at 0.05; ns, not significant.)
In previous work using Western blots of intact cells and cell-free culture media probed with an RcGTA antiserum, we showed that loss of the cckA gene in the WT strain SB1003 background results in the accumulation of RcGTA intracellularly, presumably because cell lysis is abolished (21, 35). The cckA knockout in the SBpGΔ280 background interfered with lysis because it decreased the release of pigments from cells to the level for the parental strain SBpG. Complementation of this cckA knockout strain with the WT cckA gene on a plasmid increased lysis. The release of pigments was low in the cckA knockout strain expressing the cckA H399A gene, so the absence of kinase activity coupled with the retention of phosphatase activity decreases cell lysis. This result is consistent with earlier work showing massive accumulation of the RcGTA capsid protein within cells of a derivative of the WT strain SB1003 expressing the cckA H399A gene, and the suggestion that CtrA is needed to induce the head/tail gene cluster, whereas CtrA∼P is needed for lysis (35).
In contrast to the H399A mutant, the Y589D mutant restored cell lysis to approximately the level seen with complementation using the WT cckA gene (Fig. 10). Therefore, the retention of CckA kinase activity coupled with the absence of phosphatase activity allows for a near-WT degree of cell lysis. This result is consistent with earlier work in which it was suggested that CtrA∼P is needed for cell lysis whereas CtrA is needed for the expression of head/tail genes (35).
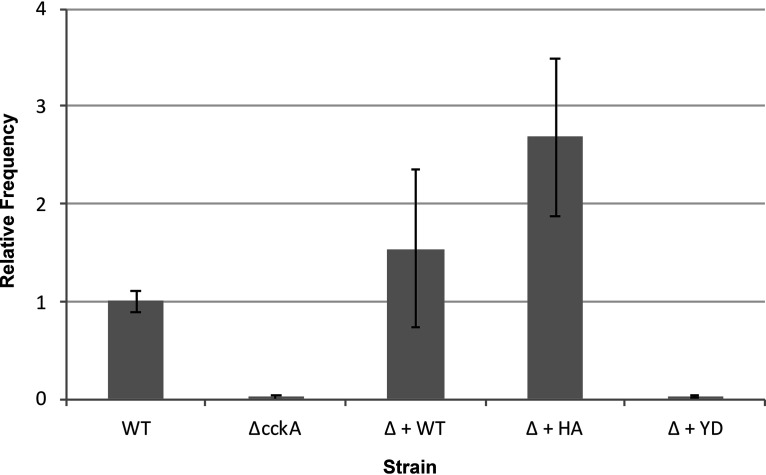
According to a model proposed by Westbye et al. (35), CtrA is needed for high levels of RcGTA particles to be produced intracellularly, whereas CtrA∼P is needed for release by cell lysis. The data in Fig. 10 show differences in cell lysis associated with the cckA allele but do not address whether RcGTA was produced and released from the strains studied. This model was addressed in a gene transfer frequency assay, which indirectly measures the relative amounts of RcGTA produced and released from gene donor cells by measuring the number of gene recipient cells, in this case transferring chromosomally encoded rifampin resistance.
As shown in Fig. 11, the cckA knockout strain produced very little functional RcGTA, and complementation of this mutant with the WT cckA gene increased the number of gene transfers. The frequency of gene transfer using the H399A mutant as the source of RcGTA might be higher than expected based on the weak lysis of this strain (Fig. 10). However, we previously found an enormous accumulation of the RcGTA capsid protein within cells of a derivative of the WT strain SB1003 expressing the cckA H399A gene (35), so we attribute the gene transfer frequency of the H399A mutant to a small number of cells lysing, with each cell producing huge amounts of RcGTA. When the Y589D mutant was used as the source of RcGTA, the gene transfer frequency was very low, on the same order of magnitude as that of the cckA knockout strain.
FIG 11.
Assay of relative gene transfer frequencies using the chromosomal marker of rifampin resistance. Abbreviations: WT, the rifampin-resistant strain SB1003; Δ, a deletion of the cckA gene (21); + WT, plasmid pRCckA, expressing the WT cckA gene; + HA, plasmid pCW104, expressing the H399A allele of cckA; + YD, plasmid pRCckA-YD, expressing the Y589D allele. The assays were performed as described elsewhere (21), and the data are normalized to the WT value (average of 516 rifampin-resistant colonies). Error bars indicate standard deviations (n = 3).
These gene transfer frequency data are supported by a Western blot of cell-free culture supernatants probed with an antiserum directed against the RcGTA capsid protein, measuring the amount of RcGTA released from cells (37). As shown in Fig. S9, the WT strain SB1003 yielded a Western blot band corresponding to the capsid protein, whereas a ΔcckA deletion mutant derived from SB1003 lacked detectable RcGTA capsid protein in the culture supernatant. The plasmid-borne WT cckA gene restored a Western blot band, as did the allele encoding H399A, whereas the Y589D protein did not restore the band.
Taking these data together, the lysis of Y589D cells (Fig. 10), the low frequency of gene transfer with Y589D mutant cultures as donors (Fig. 11), the absence of the RcGTA capsid protein in the culture media of Y589D mutant cultures (Fig. S9), and the in vitro data on the effects of this mutation on CckA phosphatase activity and the binding of c-di-GMP indicate that the binding of c-di-GMP by the CckA protein activates the phosphatase activity that is needed for CtrA-dependent production of RcGTA, whereas CtrA∼P is needed for cell lysis.
DISCUSSION
Homologues of the CckA, ChpT, and CtrA proteins are widespread in the alphaproteobacteria, but these proteins have been studied biochemically in few species. Here we show, by using an in vitro assay with purified proteins, that CckA, ChpT, and CtrA form a phosphorelay in R. capsulatus, so this bacterium now joins C. crescentus and Brucella abortus as one of the only species for which autophosphorylation and direct transfer of phosphate in this pathway have been demonstrated biochemically (38). The inclusion of c-di-GMP in the assay changed the R. capsulatus CckAΔ69 protein from a kinase to a phosphatase, converting CtrA∼P to CtrA. In previous work, it was found that in the exponential phase, CtrA appears to be predominantly nonphosphorylated and induces the expression of RcGTA structural genes indirectly by activating the transcription of gafA, which encodes a transcription factor that induces the RcGTA head-and-tail gene cluster as cultures enter the stationary phase of growth (21, 35, 39).
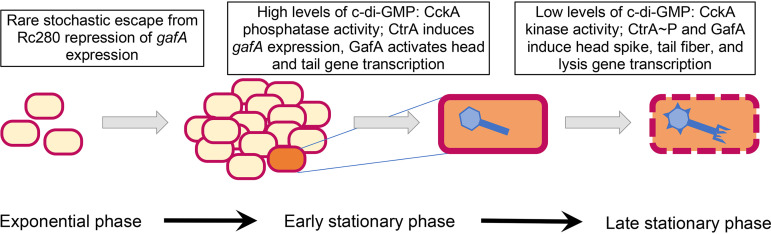
As cultures progress into the stationary phase, CtrA appears to become phosphorylated and activates the expression of the RcGTA head spike and lysis genes (35). We suggest that the level of c-di-GMP is high when cultures are in the exponential and early-stationary phases of growth, and CckA phosphatase activity maintains CtrA in the nonphosphorylated state; the c-di-GMP level decreases when cultures enter the stationary phase, and CckA kinase activity increases the concentration of CtrA∼P. A model that features the role of c-di-GMP in this growth-phase-dependent change in gene expression is shown in Fig. 12.
FIG 12.
Current model of the pathway of RcGTA induction and the role of c-di-GMP. At a low starting concentration of cells, RcGTA structural genes are not expressed during the exponential phase of growth. As the concentration of cells increases and the culture approaches the stationary phase, a small percentage of cells escape from repression of the expression of the gafA gene (encoding an alternative sigma factor) by the extracellular protein Rc280. The GafA protein activates transcription of the RcGTA head and tail structural genes, as well as feeding back to amplify the transcription of the gafA gene itself. The CckA protein acts as a phosphatase because c-di-GMP levels are high, and the CtrA protein contributes to the induction of the gafA gene, leading to massive production of RcGTA head and tail proteins. Eventually, an unknown signal decreases the concentration of c-di-GMP, which changes CckA from a phosphatase to a kinase, and with an increasing concentration of CtrA∼P, the head spike and tail fiber genes are induced, as well as lysis genes leading to the release of mature RcGTA particles. In addition to the data in this paper, the model is supported by previous publications (21, 35, 39).
The genome of R. capsulatus is predicted to encode 20 proteins that contain domains associated with the synthesis and/or degradation of c-di-GMP (23). Nine of these genes were identified in a previous transcriptomic study as having significantly reduced transcript levels in the absence of CtrA (8), and disruptions of four of those genes affected RcGTA production via changes in c-di-GMP levels (24). Together, these findings suggest that there is a feedback loop operating where CtrA regulates intracellular c-di-GMP levels by inducing the transcription of these genes, and c-di-GMP affects CtrA phosphorylation (and hence its activity) via the effect on CckA. A better understanding of the regulation of these c-di-GMP cycling enzymes and the nature of changes in c-di-GMP levels according to growth conditions and the phase of culture growth will help to provide further insight as to how this feedback system might operate.
The PAS domain protein DivL binds to CckA and inhibits kinase activity in C. crescentus (32), and genetic experiments indicate a similar role for the R. capsulatus homologue encoded by rcc00042 (35, 40). Therefore, c-di-GMP and DivL appear to have opposing effects on the CckA-ChpT-CtrA phosphorelay in the alphaproteobacteria, possibly because DivL blocks the binding of c-di-GMP to CckA. In C. crescentus, c-di-GMP also has a role in regulating ClpXP-dependent proteolysis of CtrA, and therefore, increased levels of c-di-GMP affect the cell cycle both by increasing the dephosphorylation of CtrA∼P and by increasing the degradation of the protein (41). R. capsulatus CtrA is also regulated by ClpXP (35); however, it is not known whether c-di-GMP influences CtrA protein levels in R. capsulatus.
Previous work led to a model in which nonphosphorylated CtrA is needed for the expression of RcGTA “early” genes encoding head and tail components, while CtrA∼P is required for the expression of “late” genes encoding head spike, tail fiber, and lysis proteins (35, 39). Our current results support this model, because the kinase-deficient, phosphatase-competent CckAΔ69 H399A mutant did not restore cell lysis to a ΔcckA mutant strain, whereas the phosphatase-deficient, kinase-competent CckAΔ69 Y589D mutant restored cell lysis but did not restore the release of RcGTA particles. It therefore appears that c-di-GMP, by regulating CckA kinase/phosphatase activity, controls the timing of RcGTA gene expression, resulting in the orderly maturation and release of particles.
The switch between kinase and phosphatase activity of some histidine kinases changes in response to ADP (31), and the binding of ADP in concert with c-di-GMP stimulates the phosphatase activity of C. crescentus CckA, apparently because the binding of ADP stabilizes the binding of c-di-GMP (17, 32). In our hands, the addition of ADP to a phosphorelay assay in which CtrA was phosphorylated using acetyl∼P did not detectably stimulate the phosphatase activity of the CckAΔ69 protein (Fig. 5A). We note that the degree of the effect of ADP on the kinase versus phosphatase activity of C. crescentus CckA has been reported to differ by different groups (17, 32), and our results increase the variability of the effect of ADP on these CckA activities. R. capsulatus CckA contains all the residues implicated in H-bonding to ADP in C. crescentus (see Fig. S10 in the supplemental material), indicating that there are other species-specific differences that relate to possible effects of ADP on CckA activity.
The CckAΔ69 H399A mutation abolished kinase activity but retained phosphatase activity (Fig. 7), as was also shown for C. crescentus CckA (29). As has been noted previously (42, 43), the phosphatase activity is not simply a reversal of the kinase activity. When a protein is acting as a kinase, a phosphate group is transferred from ATP to histidine upon autophosphorylation, and a covalent bond is formed between histidine and phosphate. When a protein is acting as a phosphatase, the Asp∼P bond is cleaved to form free phosphate. Although the histidine residue accelerates phosphatase activity, it is not essential for phosphatase activity.
The lessons learned from studies of the CckA-ChpT-CtrA phosphorelay of C. crescentus and related species have been valuable for understanding genome replication, morphological differentiation, and other physiological properties. However, as Brilli et al. (5) proposed, there are significant differences between the C. crescentus paradigm exhibited by members of the Caulobacterales and Rhizobiales, on the one hand, and that of species in the Rhodobacterales, on the other hand. As we showed previously, a major difference is that the C. crescentus cckA and ctrA genes are essential whereas the R. capsulatus homologues are not (8, 19), and as we show here, the effect of ADP on R. capsulatus CckAΔ69 phosphatase activity is negligible, whereas ADP significantly stimulates C. crescentus CckA phosphatase activity (17, 32).
In summary, by using His-tagged proteins, we show that c-di-GMP levels control the R. capsulatus CckA phosphatase versus kinase activity in vitro, resulting in changes in the concentration of CtrA versus CtrA∼P. Other experiments on purified proteins showed that the CckA H399A and Y589D mutant proteins differed in relative phosphatase/kinase activity and that these differences in activity correlate with differences in the binding of c-di-GMP. The effects of CckAΔ69 H399A and Y589D mutations on in vivo physiology predicted by the in vitro experiments were confirmed by the expression of nontagged, full-length cckA alleles in cell cultures where a cckA chromosomal deletion strain was complemented with WT and mutant alleles.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Escherichia coli DH5α was used for DNA manipulation and cloning (44); strain BL21(DE3) (Qiagen) was used for protein overexpression; and S17-1 lambda pir (45) was used for conjugation of plasmids. The R. capsulatus strain SBpGΔcckAΔ280 contains a chromosomally integrated mCherry fluorescent protein gene transcribed from the RcGTA head-tail gene cluster promoter (46), the cckA gene disrupted by replacement of >90% of the coding region with a kanamycin resistance cartridge (21), and a deletion of the rcc00280 gene that increases the percentage of cells that express RcGTA genes (36). E. coli was grown at 37°C in LB culture medium, supplemented as appropriate with 150 µg/ml ampicillin, 50 µg/ml kanamycin, or 10 µg/ml tetracycline. R. capsulatus was grown at 30°C in RCVm medium supplemented with 25 µg/ml kanamycin and 0.5 µg/ml tetracycline for the maintenance of plasmids (47).
Construction of mutants and plasmids.
Lists of the plasmids and primers used in this study are given in Tables S1 and S2 in the supplemental material, respectively. The broad-host-range plasmid pRK415 encoding tetracycline resistance (48) was used as the vector to mobilize full-length cckA alleles by conjugation for the complementation of R. capsulatus mutants. Plasmid pRCckA contains the WT cckA gene transcribed from the native promoter (47), and pCW104, encoding the CckA H399A protein, was constructed by site-directed mutagenesis of pRCckA as described previously (35). Plasmid pRCckA-YD, encoding the CckA Y589D protein, was constructed by site-directed mutagenesis of the cckA segment of pRCckA subcloned into pUC19 (Invitrogen) as an XbaI-to-HindII fragment, using the mutagenic primer pair CckA_Y589D-F #1 and CckA_Y589D-R #1 (Table S2). After verification of the DNA sequence, the XbaI-to-HindII fragment containing the cckA Y589D allele was transferred to pRK415 to yield pRCckA-YD.
To create a recombinant C-terminally 6His-tagged protein, the cckA gene (rcc01749) excluding the region encoding the N-terminal transmembrane region was amplified using gene-specific primers (Table S2) and was cloned as a NcoI/HindIII fragment into pET28-a. The resulting plasmid was sequence-confirmed and transformed into E. coli BL21(DE3) for protein purification. The same plasmid was also used as a template for performing site-directed mutagenesis (SDM) using the QuikChange Lightning SDM kit (Agilent Technologies) according to the manufacturer’s instructions. Mutagenesis PCRs were done using PfuUltra high-fidelity DNA polymerase and site-specific primers (Table S2) designed to create point mutations to change histidine 399 to alanine (H399A) and tyrosine 589 to aspartate (Y589D). Plasmids carrying mutations were confirmed by sequencing and transformed into E. coli BL21(DE3) for protein purification.
Purification of recombinant CckA proteins for c-di-GMP binding assays.
For protein production, overnight cultures were used to inoculate 200 ml of LB broth with kanamycin and were incubated at 30°C with shaking at 220 rpm. After 1 h, isopropyl-β-d-thiogalactopyranoside (IPTG) was added at a final concentration of 1 mM, and cells were incubated for a further 5 to 6 h. Cells were harvested by centrifugation, and cell pellets were resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 0.1% [vol/vol] Benzonase nuclease [Qiagen], 1 mg ml−1 [wt/vol] lysozyme [pH 8]) and were incubated on ice for 30 min. The suspended mixture was centrifuged at 10,000 × g for 30 min at 4°C. The supernatant was collected, mixed 4:1 (vol/vol) with Ni-nitrilotriacetic acid (Ni-NTA)–agarose slurry (Qiagen), and incubated at 4°C for 1 h. After incubation, the mixture was loaded into a column and washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole [pH 8.0]), and the recombinant proteins were eluted in four 0.5-ml aliquots of elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]). The eluted proteins were analyzed by SDS-PAGE and Coomassie brilliant blue staining. The eluted protein fractions were pooled as appropriate, dialyzed against dialysis buffer (50 mM NaH2PO4, 300 mM NaCl [pH 8]), and quantified using a Bradford protein assay (49).
c-di-GMP binding assays.
In vitro pulldown assays using streptavidin beads and biotinylated c-di-GMP were performed to assay c-di-GMP binding by CckA as described previously (50), with minor modifications. Briefly, a 20-µl reaction mixture containing the purified CckAΔTM protein (10 µM), with or without 200 pmol of biotinylated c-di-GMP, 2 µl of 10× reaction buffer (100 mM Tris, 1 M KCl, 10 mM dithiothreitol [DTT] [pH 7.5]), and 2 mM EDTA, and with or without 5 mM ADP, was prepared and incubated at room temperature for 1 h. The reaction mixture was then mixed with streptavidin beads (blocked with 5% skim milk) in 250 µl TBST (20 mM Tris, 137 mM NaCl, 0.1% Tween 20 [pH 7.5]) and was incubated at room temperature on a vortex mixer with shaking at 1,400 rpm for 1 h. The beads were collected using a magnetic strand, and the supernatant was removed. The beads were washed four to five times by adding 1 ml of TBST and incubating on the vortex mixer for 15 min at room temperature. The beads were then resuspended in 15 µl of H2O and mixed with 5 µl of 3× SDS-PAGE loading buffer.
The streptavidin bead suspensions were analyzed to detect and quantify the His-tagged proteins by Western blotting. The samples were heated at 98°C for 5 min, followed by collection of the beads on a magnetic stand, and the supernatant was collected and run on a 10% SDS-PAGE gel. The proteins were transferred to nitrocellulose membranes by electroblotting in transfer buffer (48 mM Tris base, 39 mM glycine, 20% methanol [vol/vol]). The membranes were blocked with a 5% (wt/vol) skim milk solution in TBST and incubated with the primary antibody, anti-His Tag protein (Thermo Fisher Scientific), overnight at 4°C. After a wash with TBST, membranes were incubated with the secondary antibody, peroxidase-conjugated anti-rabbit IgG (Santa Cruz Biotechnology), at room temperature for 1 h. The SuperSignal West Femto reagent kit (Thermo Fisher Scientific) was used to detect the bands by chemiluminescence, and images were captured using an Agilent ImageQuant LAS 4000 imaging system. Images were inverted and adjusted for brightness and contrast, and band intensities were quantified using ImageJ (51).
Purification of His-tagged proteins for in vitro phosphorelay experiments.
E. coli BL21(DE3) cells containing plasmids were cultivated overnight in 8 ml of medium, and these cultures were used to inoculate 300 ml of medium; the mixtures were then incubated until the cultures reached a Klett-Summerson turbidity of ∼80 Klett units (100 Klett units = ∼4 × 108 CFU/ml). At this point, IPTG was added to a final concentration of 1 mM, and incubation was continued for 3 h. After incubation, cultures were centrifuged and cell pellets frozen overnight at −20°C. Cell pellets were thawed, resuspended in 4 ml of native purification buffer (50 mM Na2HPO4, 0.5 M NaCl [pH 8.0]) supplemented with lysozyme and DNase I, incubated on ice for 10 min, and subsequently disrupted in a chilled French pressure cell (52). The resulting lysate was centrifuged at 4,000 × g for 15 min to remove cell debris, and the supernatant was retained for protein purification using column chromatography over 2 ml of Ni-NTA–agarose. After binding of the protein, the column was washed with 40 ml of native purification buffer containing 20 mM imidazole and was eluted with 5 ml of the same buffer containing 250 mM imidazole. Elution of protein was monitored by optical density at 280 nm (OD280), and fractions were pooled, diluted to a volume of 12 ml with kinase activity buffer (10 mM HEPES-KOH, 50 mM KCl, 10% glycerol, 5 mM MgCl2 [pH 8.0]) (53), and concentrated using a 10-kDa-molecular-weight-cutoff centrifugation device (Pall Corporation) at 5,000 × g for 30 min. The retained solution was then diluted in kinase buffer to a volume of 4 ml and the buffer exchange completed by four successive dilutions and concentrations over a centrifugation device (Amicon) of the appropriate molecular weight cutoff (50 kDa for CckA; 10 kDa for ChpT and CtrA), yielding samples in kinase activity buffer at a final volume of 2 ml. The concentration of purified proteins was determined by the OD at 280 nm using calculated extinction coefficients, and samples were stored at −80°C in 50-µl aliquots.
Phosphotransfer profiling and kinase/phosphatase assays.
For the evaluation of autophosphorylation and phosphate transfer in the phosphorelay, reaction mixtures were prepared in kinase activity buffer at a volume of 15 µl with the following final concentrations: 7 µM CckA, 27 µM ChpT, and 22 µM CtrA (either singly or in mixtures). The reaction was started by the addition of ATP (700 µM) or acetyl∼P (44 mM). For CckA kinase/phosphatase activity assays, a master mix was prepared consisting of 5 µM CckA, 15 µM ChpT, and 15 µM CtrA in kinase activity buffer. When required, reaction mixtures were supplemented with c-di-GMP (final concentration, 5 µM) or ADP (final concentration, 5 mM). The total volume of the reaction mixture was determined by the number of time points to be taken, with 15-µl samples taken at each time point. Phosphotransfer reactions were started by the addition of the corresponding phosphate donor (ATP or acetyl∼P), and reaction mixtures were incubated at 30°C. Reactions were stopped at the times indicated in the figures by mixing samples with SDS-PAGE loading buffer and placing them on ice.
Phos-tag PAGE.
Polyacrylamide gels (10% for CckA-only gels, 12% for gels with other proteins present) were prepared as recommended by the supplier of Phos-tag (Wako), which was included at a 50 µM concentration along with 100 mM MnCl2. SDS was not added to the gels, but it was present in the running buffer (0.4% [wt/vol] SDS, 25 mM Tris, 192 mM glycine). All gels were run for 90 to 120 min at 100 V. After electrophoresis, gels were stained with Coomassie blue in a solution of 30% methanol–10% acetic acid and were destained in the same solution lacking the dye. Gels were scanned and imaged using a Li-Cor Odyssey 9120 imaging system at a wavelength of 700 nm.
Mass spectrometry.
Samples at a concentration of 1 mg/ml were diluted 1:250 in a solution of 5% acetonitrile and 0.1% formic acid. Five microliters of this sample was then injected into a 5-mm C4 PepMap desalting column (LC Packings) connected to a Waters Xevo GS-2 QTof mass spectrometer via a NanoAquity ultraperformance liquid chromatography (UPLC) system. Samples were eluted in a 2-min gradient from 5 to 100% acetonitrile at 20 μl/min. The spectra were summed and deconvoluted using Waters’ MaxEnt algorithm.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeanette A. Johnson for expert technical assistance.
We thank the Canadian Natural Sciences and Engineering Research Council (NSERC) for financial support to J.T.B. (RGPIN 2018-03898) and A.S.L. (RGPIN 2017-04636).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wolanin PM, Thomason PA, Stock JB. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jacobs C, Domian IJ, Maddock JR, Shapiro L. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 3.Biondi EG, Reisinger SJ, Skerker JM, Arif M, Perchuk BS, Ryan KR, Laub MT. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899–904. doi: 10.1038/nature05321. [DOI] [PubMed] [Google Scholar]
- 4.Quon KC, Marczynski GT, Shapiro L. 1996. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell 84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 5.Brilli M, Fondi M, Fani R, Mengoni A, Ferri L, Bazzicalupo M, Biondi EG. 2010. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: a comparative genomic analysis. BMC Syst Biol 4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greene SE, Brilli M, Biondi EG, Komeili A. 2012. Analysis of the CtrA pathway in Magnetospirillum reveals an ancestral role in motility in alphaproteobacteria. J Bacteriol 194:2973–2986. doi: 10.1128/JB.00170-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol Microbiol 43:945–960. doi: 10.1046/j.1365-2958.2002.02777.x. [DOI] [PubMed] [Google Scholar]
- 8.Mercer RG, Callister SJ, Lipton MS, Pasa-Tolic L, Strnad H, Paces V, Beatty JT, Lang AS. 2010. Loss of the response regulator CtrA causes pleiotropic effects on gene expression but does not affect growth phase regulation in Rhodobacter capsulatus. J Bacteriol 192:2701–2710. doi: 10.1128/JB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koppenhofer S, Wang H, Scharfe M, Kaever V, Wagner-Dobler I, Tomasch J. 2019. Integrated transcriptional regulatory network of quorum sensing, replication control, and SOS response in Dinoroseobacter shibae. Front Microbiol 10:803. doi: 10.3389/fmicb.2019.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zan J, Heindl JE, Liu Y, Fuqua C, Hill RT. 2013. The CckA-ChpT-CtrA phosphorelay system is regulated by quorum sensing and controls flagellar motility in the marine sponge symbiont Ruegeria sp. KLH11. PLoS One 8:e66346. doi: 10.1371/journal.pone.0066346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsokos CG, Perchuk BS, Laub MT. 2011. A dynamic complex of signaling proteins uses polar localization to regulate cell-fate asymmetry in Caulobacter crescentus. Dev Cell 20:329–341. doi: 10.1016/j.devcel.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lori C, Ozaki S, Steiner S, Bohm R, Abel S, Dubey BN, Schirmer T, Hiller S, Jenal U. 2015. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature 523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 13.Huynh TN, Stewart V. 2011. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol Microbiol 82:275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgellis D, Kwon O, De Wulf P, Lin ECC. 1998. Signal decay through a reverse phosphorelay in the arc two-component signal transduction system. J Biol Chem 273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 15.Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281. doi: 10.1038/325279a0. [DOI] [PubMed] [Google Scholar]
- 16.Jenal U, Reinders A, Lori C. 2017. Cyclic di-GMP: second messenger extraordinaire. Nat Rev Microbiol 15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 17.Dubey BN, Lori C, Ozaki S, Fucile G, Plaza-Menacho I, Jenal U, Schirmer T. 2016. Cyclic di-GMP mediates a histidine kinase/phosphatase switch by noncovalent domain cross-linking. Sci Adv 2:e1600823. doi: 10.1126/sciadv.1600823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaczmarczyk A, Hempel AM, von Arx C, Bohm R, Dubey BN, Nesper J, Schirmer T, Hiller S, Jenal U. 2020. Precise timing of transcription by c-di-GMP coordinates cell cycle and morphogenesis in Caulobacter. Nat Commun 11:816. doi: 10.1038/s41467-020-14585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang AS, Beatty JT. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc Natl Acad Sci U S A 97:859–864. doi: 10.1073/pnas.97.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang AS, Westbye AB, Beatty JT. 2017. The distribution, evolution, and roles of gene transfer agents in prokaryotic genetic exchange. Annu Rev Virol 4:87–104. doi: 10.1146/annurev-virology-101416-041624. [DOI] [PubMed] [Google Scholar]
- 21.Mercer RG, Quinlan M, Rose AR, Noll S, Beatty JT, Lang AS. 2012. Regulatory systems controlling motility and gene transfer agent production and release in Rhodobacter capsulatus. FEMS Microbiol Lett 331:53–62. doi: 10.1111/j.1574-6968.2012.02553.x. [DOI] [PubMed] [Google Scholar]
- 22.Lang AS, Beatty JT. 2002. A bacterial signal transduction system controls genetic exchange and motility. J Bacteriol 184:913–918. doi: 10.1128/jb.184.4.913-918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strnad H, Lapidus A, Paces J, Ulbrich P, Vlcek C, Paces V, Haselkorn R. 2010. Complete genome sequence of the photosynthetic purple nonsulfur bacterium Rhodobacter capsulatus SB 1003. J Bacteriol 192:3545–3546. doi: 10.1128/JB.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pallegar P, Pena-Castillo L, Langille E, Gomelsky M, Lang AS. 2019. Cyclic di-GMP-mediated regulation of gene transfer and motility in Rhodobacter capsulatus. J Bacteriol 202:e00554-19. doi: 10.1128/JB.00554-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H, Ziesche L, Frank O, Michael V, Martin M, Petersen J, Schulz S, Wagner-Dobler I, Tomasch J. 2014. The CtrA phosphorelay integrates differentiation and communication in the marine alphaproteobacterium Dinoroseobacter shibae. BMC Genomics 15:130. doi: 10.1186/1471-2164-15-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou B, Schrader JM, Kalogeraki VS, Abeliuk E, Dinh CB, Pham JQ, Cui ZZ, Dill DL, McAdams HH, Shapiro L. 2015. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet 11:e1004831. doi: 10.1371/journal.pgen.1004831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinoshita E, Kinoshita-Kikuta E. 2011. Improved Phos-tag SDS-PAGE under neutral pH conditions for advanced protein phosphorylation profiling. Proteomics 11:319–323. doi: 10.1002/pmic.201000472. [DOI] [PubMed] [Google Scholar]
- 28.Lukat GS, McCleary WR, Stock AM, Stock JB. 1992. Phosphorylation of bacterial response regulator proteins by low molecular weight phospho-donors. Proc Natl Acad Sci U S A 89:718–722. doi: 10.1073/pnas.89.2.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. 2009. Dynamics of two phosphorelays controlling cell cycle progression in Caulobacter crescentus. J Bacteriol 191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubey BN, Agustoni E, Böhm R, Kaczmarczyk A, Mangia F, von Arx C, Jenal U, Hiller S, Plaza-Menacho I, Schirmer T. 2020. Hybrid histidine kinase activation by cyclic di-GMP–mediated domain liberation. Proc Natl Acad Sci U S A 117:1000–1008. doi: 10.1073/pnas.1911427117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mideros-Mora C, Miguel-Romero L, Felipe-Ruiz A, Casino P, Marina A. 2020. Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases. Nat Commun 11:769. doi: 10.1038/s41467-020-14540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mann TH, Shapiro L. 2018. Integration of cell cycle signals by multi-PAS domain kinases. Proc Natl Acad Sci U S A 115:E7166–E7173. doi: 10.1073/pnas.1808543115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Heindl JE, Fuqua C. 2013. Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens. PLoS One 8:e56682. doi: 10.1371/journal.pone.0056682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dye KJ, Yang ZM. 2020. Cyclic-di-GMP and ADP bind to separate domains of PilB as mutual allosteric effectors. Biochem J 477:213–226. doi: 10.1042/BCJ20190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westbye AB, Kater L, Wiesmann C, Ding H, Yip CK, Beatty JT. 2018. The protease ClpXP and the PAS domain protein DivL regulate CtrA and gene transfer agent production in Rhodobacter capsulatus. Appl Environ Microbiol 84:e00275-18. doi: 10.1128/AEM.00275-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding H, Grull MP, Mulligan ME, Lang AS, Beatty JT. 2019. Induction of Rhodobacter capsulatus gene transfer agent gene expression is a bistable stochastic process repressed by an extracellular calcium-binding RTX protein homologue. J Bacteriol 201:e00430-19. doi: 10.1128/JB.00430-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fu YY, MacLeod DM, Rivkin RB, Chen F, Buchan A, Lang AS. 2010. High diversity of Rhodobacterales in the subarctic North Atlantic Ocean and gene transfer agent protein expression in isolated strains. Aquat Microb Ecol 59:283–293. doi: 10.3354/ame01398. [DOI] [Google Scholar]
- 38.Willett JW, Herrou J, Briegel A, Rotskoff G, Crosson S. 2015. Structural asymmetry in a conserved signaling system that regulates division, replication, and virulence of an intracellular pathogen. Proc Natl Acad Sci U S A 112:E3709–E3718. doi: 10.1073/pnas.1503118112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fogg PCM 2019. Identification and characterization of a direct activator of a gene transfer agent. Nat Commun 10:595. doi: 10.1038/s41467-019-08526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Childers WS, Xu Q, Mann TH, Mathews II, Blair JA, Deacon AM, Shapiro L. 2014. Cell fate regulation governed by a repurposed bacterial histidine kinase. PLoS Biol 12:e1001979. doi: 10.1371/journal.pbio.1001979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev 23:93–104. doi: 10.1101/gad.502409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsing W, Silhavy TJ. 1997. Function of conserved histidine-243 in phosphatase activity of EnvZ, the sensor for porin osmoregulation in Escherichia coli. J Bacteriol 179:3729–3735. doi: 10.1128/jb.179.11.3729-3735.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casino P, Rubio V, Marina A. 2010. The mechanism of signal transduction by two-component systems. Curr Opin Struct Biol 20:763–771. doi: 10.1016/j.sbi.2010.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic-engineering—transposon mutagenesis in gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 46.Fogg PCM, Westbye AB, Beatty JT. 2012. One for all or all for one: heterogeneous expression and host cell lysis are key to gene transfer agent activity in Rhodobacter capsulatus. PLoS One 7:e43772. doi: 10.1371/journal.pone.0043772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westbye AB, Leung MM, Florizone SM, Taylor TA, Johnson JA, Fogg PC, Beatty JT. 2013. Phosphate concentration and the putative sensor kinase protein CckA modulate cell lysis and release of the Rhodobacter capsulatus gene transfer agent. J Bacteriol 195:5025–5040. doi: 10.1128/JB.00669-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keen NT, Tamaki S, Kobayashi D, Trollinger D. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 49.Bradford MM 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 50.Chambers JR, Sauer K. 2017. Detection of cyclic di-GMP binding proteins utilizing a biotinylated cyclic di-GMP pull-down assay. Methods Mol Biol 1657:317–329. doi: 10.1007/978-1-4939-7240-1_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuchinski KS, Brimacombe CA, Westbye AB, Ding H, Beatty JT. 2016. The SOS response master regulator LexA regulates the gene transfer agent of Rhodobacter capsulatus and represses transcription of the signal transduction protein CckA. J Bacteriol 198:1137–1148. doi: 10.1128/JB.00839-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.