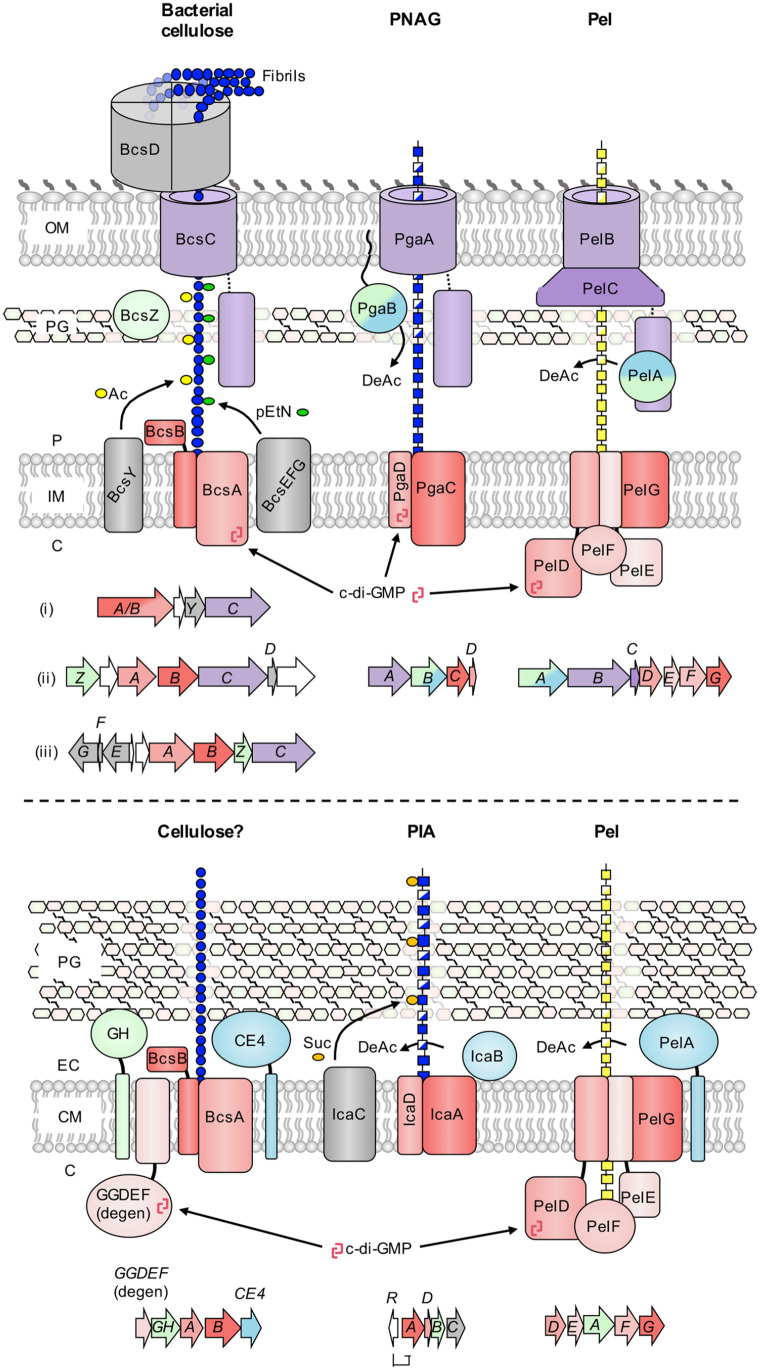
Figure 1.
Biosynthetic models for synthase-dependent polysaccharides in Gram-negative and Gram-positive bacteria. (top) Schematics for the biosynthesis of bacterial cellulose, poly-β-1,6-N-acetyl-glucosamine (PNAG), and the Pel polysaccharide in Gram-negative bacteria. Representative genetic loci are shown below each model and are drawn to scale. Three representative gene clusters are shown for bacterial cellulose due to the diversity of these operons: (i) Komagataeibacter xylinus E25 locus one, (ii) Komagataeibacter xylinus E25 locus two and (iii) Salmonella enterica serovar Typhimurium. The PNAG operon is from Escherichia coli K-12 substr. MG1655, and the Pel operon is from Pseudomonas aeruginosa PAO1. (bottom) Schematics for the biosynthesis of bacterial cellulose, polysaccharide intercellular adhesin (PIA), and the Pel polysaccharide in Gram-positive bacteria. Representative genetic loci are shown below each model and are drawn to scale. The bacterial cellulose operon is from Clostridium botulinum ATCC 3502, the PIA operon is from Staphylococcus epidermidis RP62A, and the Pel operon is from Bacillus cereus ATCC 10987. Conserved functionalities across synthase systems are coloured as follows: red, synthase components (polymerization, inner membrane transport, c-di-GMP binding); purple, tetratricopeptide-repeat and β-barrel (outer membrane export), blue, deacetylase; green, glycoside hydrolase. Variable accessory components for each system are depicted in gray. C, cytoplasm; IM, inner membrane; P, periplasm; OM, outer membrane; PG, peptidoglycan; CM, cytoplasmic membrane; EC, extracellular space; Ac, acetyl group; pEtN, phosphoethanolamine; DeAc, deacetylation; c-di-GMP, cyclic-3′,5′-dimeric guanosine monophosphate; Suc, succinyl group, GH, predicted glycoside hydrolase; CE4, predicted carbohydrate esterase family 4 deacetylase; GGDEF (degen), degenerate diguanylate cyclase with intact inhibitory site.