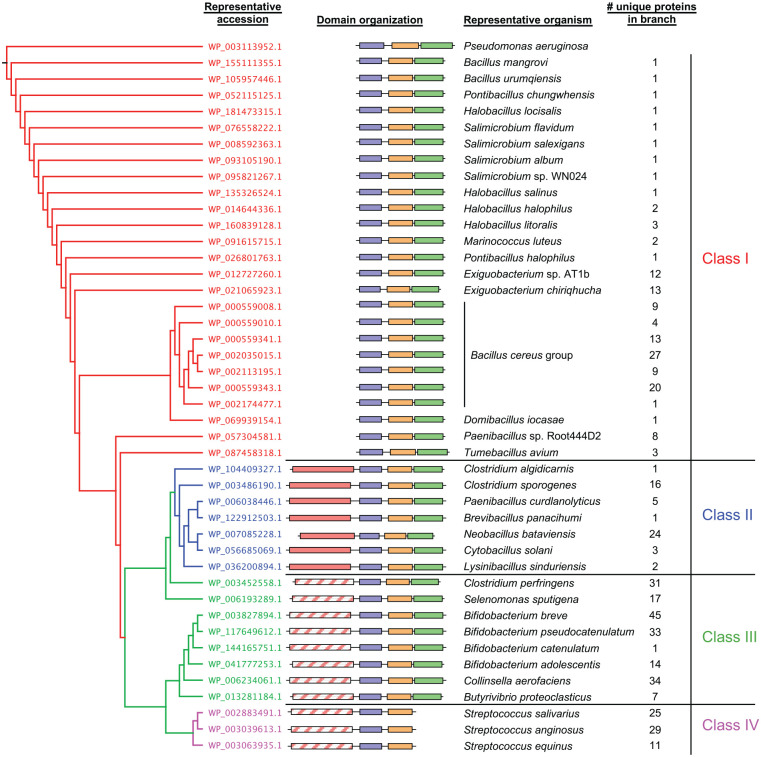
Figure 3.
Gram-positive PelD orthologs cluster into 4 different classes based on their predicted domain architectures. (left) Phylogenetic tree generated from multiple sequence alignment of PelD protein sequences. Terminal branches with many closely clustered leaves were collapsed for ease of presentation, and 1 representative PelD ortholog from each cluster is shown. Each branch is coloured according to the PelD domain architecture class, shown at the far right. (middle) Schematic representations of Gram-positive PelD domain organizations corresponding to the protein accession numbers from the phylogenetic tree. Individual domains are represented as boxes and are drawn to scale. (right) Representative bacterial species from the corresponding branch of the phylogenetic tree whose PelD domain organization is schematically depicted. The number of leaves present in each collapsed branch of the phylogenetic tree is indicated. DUF4118, domain of unknown function containing 4 predicted transmembrane helices; GAF, cGMP-specific phosphodiesterases, adenylyl cyclases and FhlA domain; GGDEF, diguanylate cyclase domain; SDR, short chain dehydrogenase/reductase domain. Protein domains are coloured by predicted function as follows: purple, DUF4118; orange, GAF; green, degenerate GGDEF; red, SDR; hatched red, degenerate SDR.