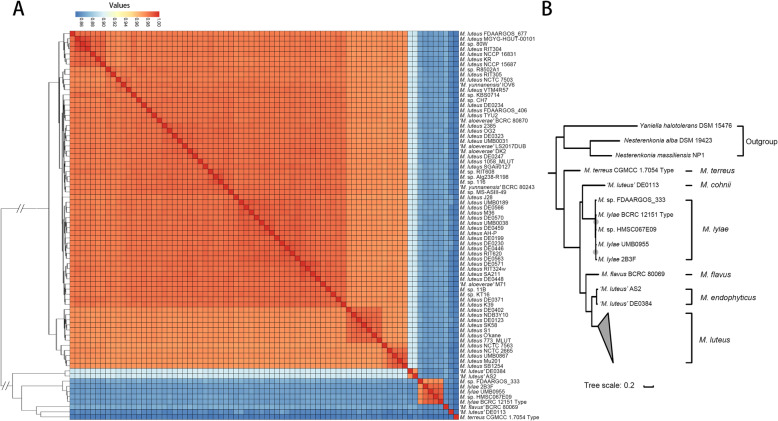
Fig. 1.
Pairwise genome-wide ANI values and maximum-likelihood phylogeny of the genus Micrococcus. a Heatmap of average nucleotide identities between Micrococcus strains used in this study. ANI values calculated by the pyani are color-coded according to the provided scale bar. b Maximum-likelihood phylogeny generated from concatenated 174 single-copy core genes with no evidence of recombination from PhiPack of the 76 Micrococcus strains, including three outgroup strains. Bootstrap values less than 90% are shown by gray dots at the nodes. The scale bar indicates 20% sequence divergence