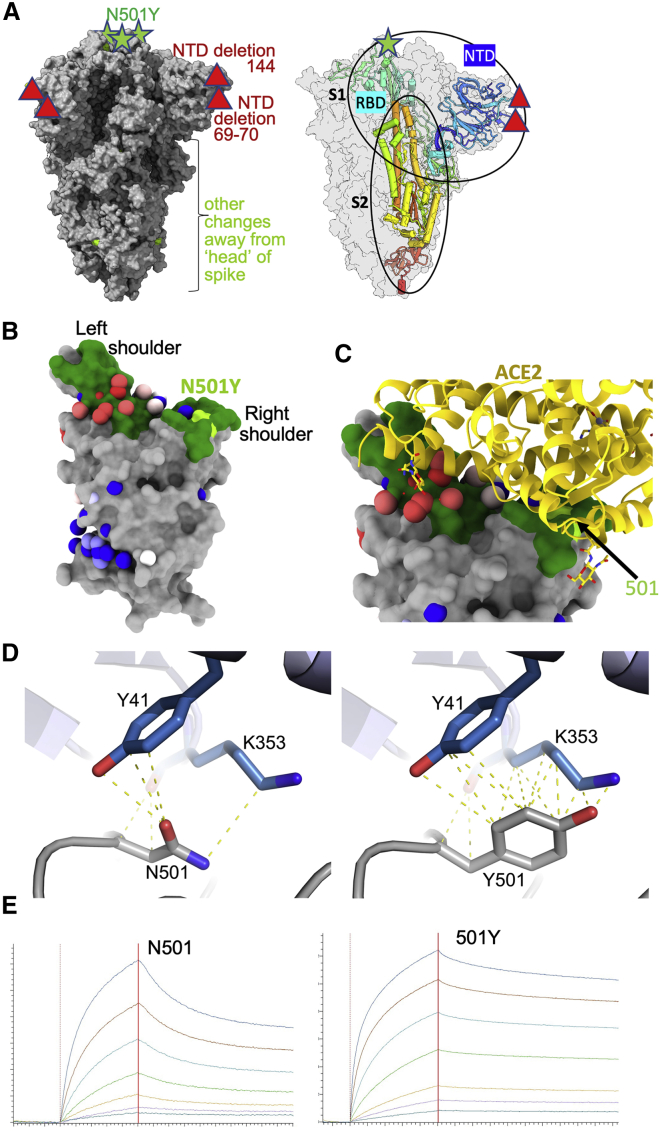
Figure 1.
The B.1.1.7 variant spike protein and effect on ACE binding of the N501Y mutation
(A) The SARS-CoV-2 spike trimer is depicted as a gray surface with mutations highlighted in yellow-green or with symbols. The RBD N501Y and the NTD 144 and 69–70 deletions are highlighted with green stars and red triangles, respectively. On the right, a protomer is highlighted as a colored ribbon within the transparent gray spike surface, illustrating its topology and marking key domains.
(B) The RBD “torso” analogy. The RBD is represented as a gray surface with the ACE2 receptor binding site in dark green. Binding sites for the panel of antibodies (Dejnirattisai et al., 2021) on which this study draws are represented by spheres. The spheres represent the point at which placing spherical antibodies would optimally predict the BLI competition data and are colored according to their neutralization, from red (potent) to blue (non-neutralizing). The position of the B.1.1.7 N501Y mutation in the RBD is highlighted in light green toward the right shoulder.
(C) Proximity of ACE2 to N501Y. The RBD is depicted as in (B) with ACE2 bound (in yellow cartoon format) with glycosylation drawn as sticks.
(D) Left panel: interactions of N501 of WT RBD with residues Y41 and K353. The structure shown is the complex of N501 RBD with ACE2 determined by X-ray crystallography (PDB ID 6M0J, Lan et al., 2020). When the 501 is mutated to a tyrosine with the conformation seen in the N501Y RBD-269 Fab complex (right panel), Y501 makes T-shaped ring stacking interactions with Y41 and more hydrophobic contacts with K353 of ACE2 (note there are minor clashes of the side chain of Y501 to the end of the K353 side chain, which has ample room to adjust to optimize interactions).
(E) BLI plots for WT (left) and N501Y (right) RBDs binding to ACE2. A titration series is shown for each (see STAR Methods). Note the much slower off rate for the mutant.