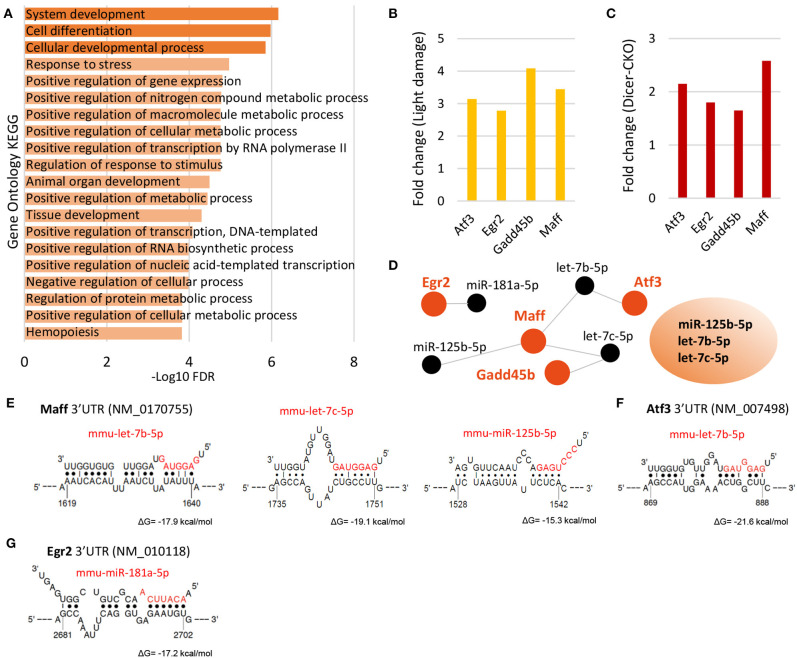
Figure 6.
Upregulated genes after light damage and after Dicer deletion and their potential miRNA regulators. (A) Gene ontology of the top 20 genes upregulated the most after Dicer deletion and light damage (LD), FDR: false discovery rate. (B,C) Top four genes upregulated after light damage (B, fold change of log2 relative expression of light damage vs. controls, five and six biological replicates, respectively, one technical replicate per condition) or Dicer deletion (C, fold change of log2 counts per million of wild type or Dicer-deleted MG, 20 and 14 biological replicates, respectively, 1 technical replicate per condition). (D) DIANA-TarBase predicted miRNAs regulating the top four genes (Atf3, Egr2, Gadd45b, and Maff ) upregulated in the Dicer-cKO and after light damage and miRNAs found to be regulating the top genes after LD and the top 4 genes in the light damage/Dicer-cKO dataset i.e., miR-125b-5p, let-7c-5p, and let-7b-5p. (E–H) Binding sites of identified miRNAs from D (DIANA-TarBase) in the 3′UTRs of Maff (E), Atf3 (F), and Egr2 (G) using STarMirDB. The seed sequences of the particular miRNA are shown in red. Dots represent complimentary base pairing. ΔG indicates the structural accessibility at the target site.