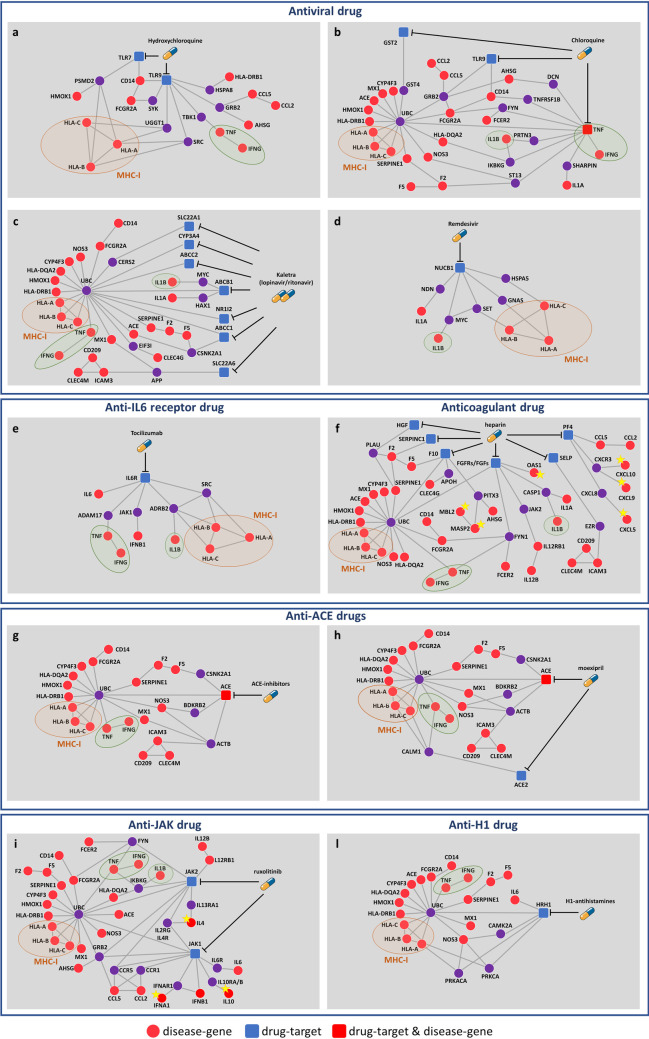
Fig 7. Mechanism-of-action of anti-SARS-CoV repurposable drugs.
The subnetworks show the inferred mechanism-of-action for: antiviral drugs (A-D), tocilizumab (E), heparin (F), ACE-inhibitors (G-H), ruxolitinib (I), and H1-antistamines (L). Each subnetwork was designed to point out the shortest paths from drug targets and SARS disease genes in the human interactome. In each subnetwork, disease genes specifically targeted by each drug are marked with a yellow star; major histocompatibility complex of class I (MCH-I) and pro-inflammatory cytokines shared by all drug networks are marked with an orange and a green circle, respectively. Legend: red circles refer to SARS disease genes, blue squares refer to drug targets, red squares refer to SARS disease genes that are also drug targets, violet circles refer to the first nearest neighbors (that are not disease genes) of the drug targets in the human interactome. Anti-ACE drugs of panel (G) refer to enalapril, trandolapril, fosinopril, benazepril, cilazapril, zofenopril, spirapril, rescinnamine, and quinapril. H1-anistamines of panel (L) refer to fexofenadine, levocetirizine, desloratadine, clemastine, cetirizine.