Figure 1.
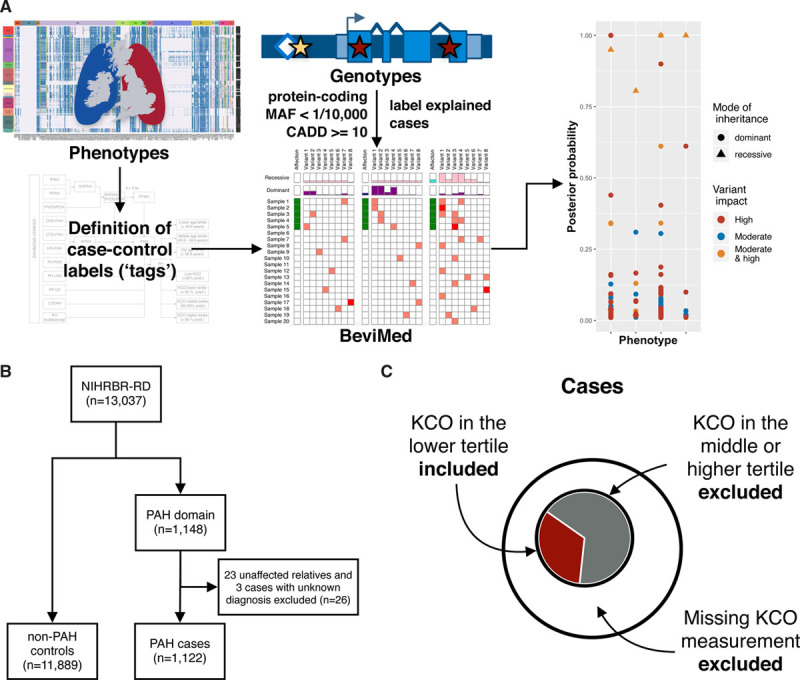
Design of the genetic association study. A, Overview of the analytical approach. Using deep phenotyping, data tags were assigned to patients who shared phenotypic features. Rare sequence variants, called from whole-genome sequencing data, were filtered, and explained cases were labeled. BeviMed was applied to a set of unrelated individuals to estimate the posterior probability of gene-tag associations. B, Consort diagram summarizing the size of the study cohort. C, Schematic representation of the definition of cases, exemplified by the transfer coefficient for carbon monoxide (KCO) lower tertile tag. Cases were defined as individuals carrying a particular tag, whereas patients with missing information or those without a tag were removed from the gene-tag association testing. Individuals from non-pulmonary arterial hypertension (PAH) domains served as controls. CADD indicates combined annotation dependent depletion; and MAF, minor allele frequency.
