Abstract
The evolution of venom resistance through coevolutionary chemical arms races has arisen multiple times throughout animalia. Prior documentation of resistance to snake venom α-neurotoxins consists of the N-glycosylation motif or the hypothesized introduction of arginine at positions 187 at the α-1 nicotinic acetylcholine receptor orthosteric site. However, no further studies have investigated the possibility of other potential forms of resistance. Using a biolayer interferometry assay, we first confirm that the previously hypothesized resistance conferred by arginine at position 187 in the honey badger does reduce binding to α-neurotoxins, which has never been functionally tested. We further discovered a novel form of α-neurotoxin resistance conferred by charge reversal mutations, whereby a negatively charged amino acid is replaced by the positively charged amino acid lysine. As venom α-neurotoxins have evolved strong positive charges on their surface to facilitate binding to the negatively charged α-1 orthosteric site, these mutations result in a positive charge/positive charge interaction electrostatically repelling the α-neurotoxins. Such a novel mechanism for resistance has gone completely undiscovered, yet this form of resistance has convergently evolved at least 10 times within snakes. These coevolutionary innovations seem to have arisen through convergent phenotypes to ultimately evolve a similar biophysical mechanism of resistance across snakes.
Keywords: resistance, alpha-neurotoxins, nicotinic acetylcholine receptors, venom, elapidae
1. Introduction
Antagonistic interactions, such as predator–prey relationships, are drivers of evolutionary novelty in the coevolution of many organisms, with reciprocal selection pressures leading to genotypic changes [1,2]. This reciprocal evolution allows positive selection pressures to drive defensive and offensive adaptations in both predators and prey. A prime example of this is the evolution of venom which leads to the development of resistance in an opposing predator or prey [3].
A prominently evolved form of resistance is seen within the muscle-type α-1 nicotinic acetylcholine receptor (nAChR) of some animals. The presence of specific amino acids reduces the relative sensitivity to neurotoxins that target the nAChRs (α-neurotoxins) found in some snake venoms [4–9]. The acetylcholine binding region (orthosteric site) is located around amino acid positions 187–200 [10] of the α-1 nAChR subunit, with positions 187, 189, 190, 191, 194 and 195 having been identified, to varying degrees, to play a role in α-neurotoxin binding [9,11–13].
Biochemically distinct residue changes at positions 187 and 189, thought to confer resistance to α-neurotoxins due to steric inhibition, had been found in mammals which predate upon neurotoxic snakes, such as the honey badger (Mellivora capensis), hedgehog (Erinaceus spp.), mongoose (Herpestes ichneumon) and wild pig (Sus scrofa) [4,7–9], as well as within cobras (Naja) as autoresistance to their own venom [4–6,14]. First discovered in the mongoose, a major predator of neurotoxic cobras, was a steric hindrance form of resistance conferred by the evolution of N-glycosylation at position 187 [4,7]. This same form of resistance, but at position 189, was later found in elapid snakes thought to be as autoresistance to their own neurotoxins [5,6,15]. Other minor forms of resistance thought to act synergistically with these primary forms are the substitutions at position 189 from non-polar aromatic phenylalanine (F) to non-polar aliphatic isoleucine (I) or leucine (L) [4]. However, this reduced binding is only thought to occur when both 187 and 189 (aromatic subsite) ancestral amino acids are substituted out [9], thus acting in a synergistic manner rather than directly driving the resistance. Another synergistic contribution to resistance are mutations at the proline (P) subsite (residues 194P and 197P) which were shown to cause a significant binding decrease toward α-bunagrotoxin when these prolines were substituted out [9]. It was suggested that the substitutions at the proline subsite play a critical role in α-neurotoxin binding while substitutions to the aromatic subsite might determine the specificity of toxin resistance [9].
Another key biochemical change thought to confer resistance is that of the positively charged arginine (R) which has convergently evolved three times in mammals at position 187 from the ancestral aromatic tryptophan (W) in honey badgers, hedgehogs and wild pigs [4,15]. The mechanism by which 187R is suspected to cause the significant reduction in binding to positively charged α-neurotoxins such as three-finger toxins (3FTxs) [16] has been postulated to be through electrostatic repulsion [4,11] or, more recently, hypothesized as being due instead to steric hinderance by the introduction of this bulky amino acid [17].
The N-glycosylation at positions 187 (mongoose) and 189 (cobra) have been tested and confirmed to confer resistance [5–7,9]. By contrast, the evolution of 187R in the honey badger has only been hypothesized to confer resistance to α-neurotoxins [4], due to them being predators of cobras (Naja) and based on natural history observations of them surviving cobra bites [4,18]. However, this hypothesis has never been functionally tested. Thus, it remains unclear if the proposed resistance is through the positively charged R causing electrostatic repulsion of the equally positively charged α-neurotoxins from cobras.
Until recently, the 187R residue change was the only positively charged substitution seen in any sequenced nAChR orthosteric site [4]. However, a comprehensive examination of published nAChR sequences across the animal kingdom [15] revealed that within Serpentes (an assessment of 76 different snake species), the positively charged amino acid lysine (K) has convergently evolved on at least 10 separate occasions by replacing a negatively charged amino acid; aspartic acid (D) or glutamic acid (E) at positions 191 or 195 (with 195 being hypervariable in having the negative amino acid). The ancestral negatively charged D and E would be highly susceptible to binding from the positively charged α-neurotoxins from snakes [19,20] especially since these positions have been shown to be crucial α-neurotoxin binding sites [9,11–13]. Indeed, the selection pressure exerted by these negatively charged sites has resulted in positively charged molecular surfaces being a general characteristic of neurotoxins binding to this site as the opposite-charge interactions would facilitate binding. Therefore, we hypothesize that evolving a positively charged residue in their place would likely cause electrostatic repulsion of the positively charged residues to the positively charged α-neurotoxins.
We first set out to functionally test and confirm if the positively charged 187R of the honey badger does in fact induce resistance/reduced susceptibility toward α-neurotoxins. We then tested our hypothesis that these charge reversals may confer resistance to α-neurotoxins through a novel charge reversal electrostatic form of repulsion.
We tested these by assessing the binding of venoms rich in α-neurotoxins from sympatric and allopatric snake species to native and site-specific amino acid point mutants of the nAChR orthosteric sequences that contain positively charged substitutions from the honey badger (M. capensis) and snakes (Atractaspis bibronii/A. microlepidota, Pseudaspis cana and Python bivittatus). These snake species were chosen based on them having critical K mutations at key positions and also being likely prey of sympatric elapid species. By further testing sympatric and allopatric snake venoms, we can also assess if any resistance is likely caused by local adaptation due to coevolution. To assess these, we used a biolayer interferometry (BLI) assay platform that has been previously validated to measure α-neurotoxic binding to nAChR orthosteric mimotopes (a small amino acid sequence representative of part of the epitope) [21–23].
2. Results and discussion
(a). Assessing the arginine (R) resistance
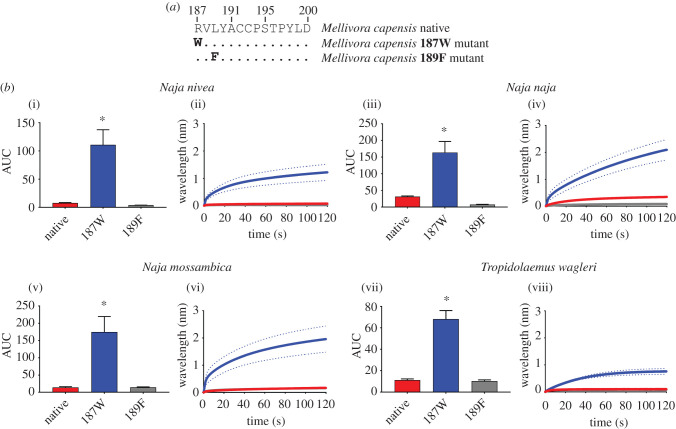
The honey badger differs from the ancestral mammalian state at two positions—187R in place of the ancestral tryptophan (W) (the denotation of ancestral changes will be displayed as W187R throughout the manuscript) and leucine (L) in place of the ancestral phenylalanine (F) at 189 (L189F). We tested the influence of these changes to the native honey badger sequence, as well as mutants with one amino acid changed back to the ancestral state (figure 1a). As expected, the native state was not bound by the three sympatric cobra venoms tested, or the α-neurotoxic venom of the viper Tropidolaemus wagleri used as an allopatric outgroup to test for generalized resistance to α-neurotoxins by the honey badger. The 189F mutant did not show any binding increase, suggesting that this change in the honey badger is not responsible for the resistance toward cobra venoms. By contrast, changing the derived 187R back to the ancestral 187W resulted in a strong increase in the binding by the venoms (figure 1b). Thus, our data suggest that the resistance is solely conferred by 187R rather than both aromatic subsite positions. The resistance to both sympatric and allopatric α-neurotoxic snake venoms suggests that although the coevolutionary selection pressures are likely due to the honey badger predating on cobras, the reduced susceptibility is also present across the allopatric α-neurotoxin types (T. wagleri) signifying similar binding mechanisms of these toxins, thus reinforcing the paradigm that sensitivities in the target provides the selection pressure for the evolution of the toxin. Therefore, while the two types of neurotoxic snake venoms tested evolved independently, their convergent evolution for the same target resulted in convergent biochemical mechanisms. The 187R has convergently evolved in some caecilians [15] that are sympatric and potential prey items for the South American members of the Elapidae snake genus Micrurus and thus may represent another independent evolution of this form of resistance.
Figure 1.
The effects of venom from four Naja species against the honey badger (Mellivora capensis) native and mutant mimotope sequences. (a) Amino acid sequences of native and mutant mimotopes. (b) Bar graphs represent the mean area under the curve (AUC) values of the adjacent curve graphs. Curve graphs show the mean wavelength (nm) shift in light with increased binding of venoms over a 120 s association phase. Each venom was tested in triplicate (n = 3). Error bars on all graphs represent the SEM. AUC values were statistically analysed using a one-way ANOVA with a Dunnett's multiple comparisons post hoc test comparing to the native mimotope. Statistical significance is indicated by an asterisk above the corresponding bar with a significance threshold of p < 0.01. All raw data and statistical analyses outputs can be found in electronic supplementary material, data S1. (Online version in colour.)
(b). Assessing the charge reversal lysine (K) resistance
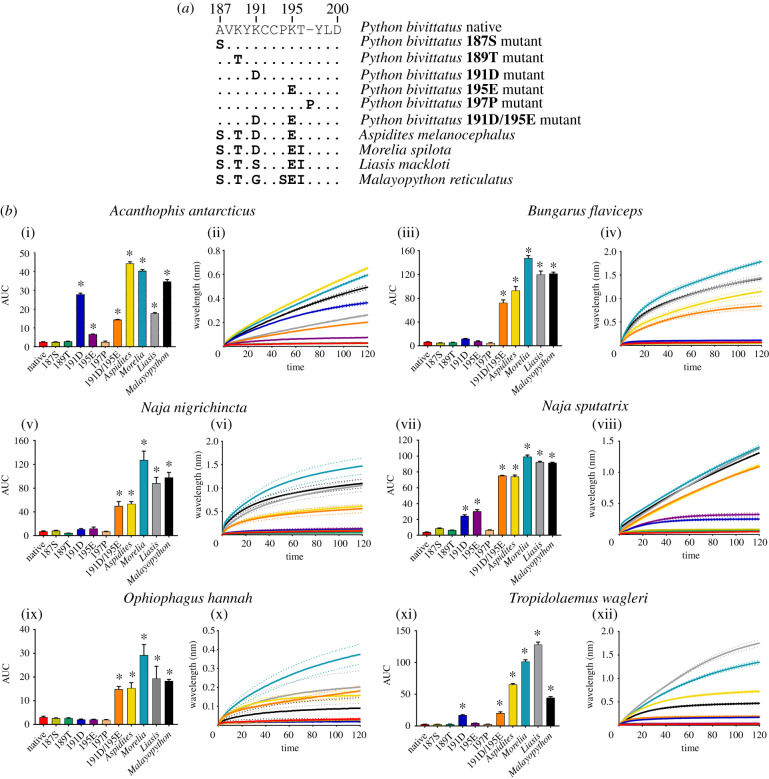
To assess the lysine form of resistance, we first tested the Burmese python (Python bivittatus), a slow-moving, predominantly terrestrial snake which when young would be vulnerable to predation by sympatric snake-eating neurotoxic venomous snakes such as cobras (Naja spp.), kraits (Bungarus spp.) and king cobras (Ophiophagus spp.). The Burmese python α-1 nAChR orthosteric site has 189K, 191K and 195K positions (figures 2 and 3). The mutations at 191 and 195 are particularly significant as they represent charge reversals, as the positively charged K replaces the negatively charged amino acids found in other pythons at position 191 (aspartic acid [D]), and position 195 (glutamic acid [E]) (figure 2). We tested both native and mutant orthosteric site sequences of P. bivittatus as well as the native sequences of closely related Pythonidae representatives: Aspidites melanocephalus, Liasis mackloti, Morelia spilota and Malayopython reticulatus, which all lack the K mutations present in P. bivittatus (figure 3a). First, the 187S mutant was used to confirm that the uncommon 187A of the P. bivittatus sequence does not induce α-neurotoxin resistance. Python bivittatus has also lost the 197P, thus this mutant was tested to assess if the removal of the 197P interferes with α-neurotoxin binding since the removal of one or both 194P/197P (the proline subsite) in mammal assays significantly reduced α-bungarotoxin binding [9]. Neither of these two changes increased binding (figure 3b). Further, there was no change in binding when the 189K was substituted out; however, changing the 191K or 195K together simultaneously increases binding across all venoms tested seeming to conform the majority of the resistance. However, it appears that substitutions of all three K significantly increase the binding more than the 191K and 195K changes alone (figure 3b). Further, these results also suggest that the charged residues might work synergistically to reduce toxin susceptibility since there was an increase when both 191K and 195K were removed (figure 3a). These positions together may induce an intensity effect by where the sum of their charge is greater at α-neurotoxin repulsion.
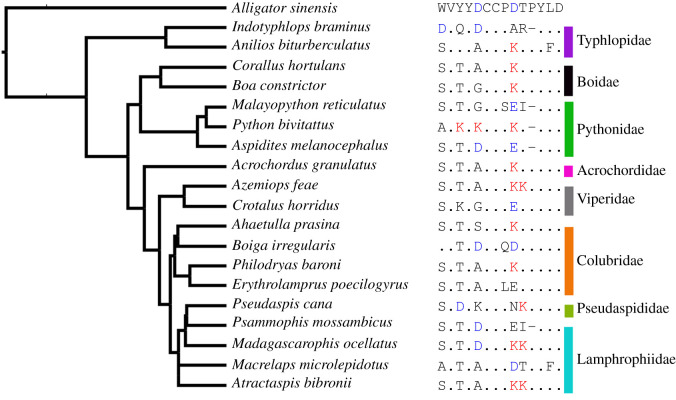
Figure 2.
Sites of positively charged lysine (K) and negatively charged aspartic acid (D) and glutamic acid (E) within serpents. The phylogenetic tree was constructed from the consensus of Timetree.org and amended based upon a previously constructed comprehensive phylogeny [15]. This phylogeny is a smaller abridged version of the nAChR sequences tree of Khan et al. [15]. The ancestral sequence (Alligator sinensis) displays all corresponding amino acid letter abbreviations with all other species sequence similarities noted as a dot except for the ancestral aspartic acid (D). Positively charged lysine (K) is highlighted in red while both negatively charged aspartic acid (D) and glutamic acid (E) are noted in blue. Deviations from the ancestral amino acids are in black. (Online version in colour.)
Figure 3.
The effects of venom from six elapid species against the Python bivittatus (native and mutants) and other Pythonidae representative mimotope sequences. (a) Amino acid sequences of native and mutant mimotopes. (b) Bar graphs represent the mean area under the curve (AUC) values of the adjacent curve graphs. Curve graphs show the mean wavelength (nm) shift in light with increased binding of venoms over a 120 s association phase. Each venom was tested in triplicate (n = 3). Error bars on all graphs represent the SEM. AUC values were statistically analysed using a one-way ANOVA with a Dunnett's multiple comparisons post hoc test comparing to the native mimotope. A statistical significance is annotated by * above the corresponding bar with a significance threshold of p < 0.01. All raw data and statistical analyses outputs can be found in electronic supplementary material, data S2. (Online version in colour.)
From an evolutionary perspective, P. bivittatus (and other members of the Python genus) are slow moving (usually due to their large girth) and predominantly terrestrial snakes [24] that are potential prey for many species of African and Asian elapids, particularly neonate and juvenile stages. Thus, they might be under a high selection pressure to evolve resistance toward α-neurotoxins. Conversely, Malayopython reticulatus showed no reduced susceptibility; however, there may not be a sufficient resistance selection pressure since as juveniles they mostly occupy the arboreal niche [24] which is devoid of elapid snakes. By adulthood Malayopython reticulatus are very large (the largest of all living snakes at up to 10 m [24]) and thus not likely to be prey. Morelia spilota are also predominantly/frequently arboreal [25], while Liasis mackloti is deemed a semi-aquatic species, habitually entering water [24,26]. However, similarly to Aspidites melanocephalus, they both inhabit geographical regions that are mostly lacking in neurotoxic snake-eating venomous snakes, and thus not under a strong selection pressure to evolve resistance.
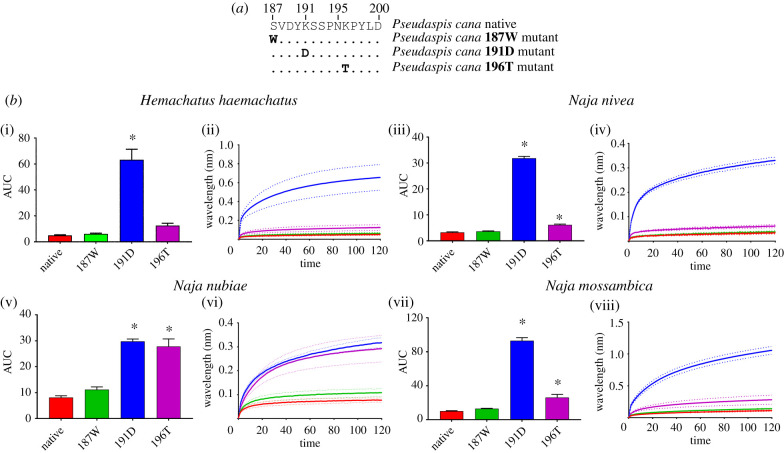
Pseudaspis cana has evolved a charge reversal D191K, and with the introduction of an additional 196K which does not represent a charge reversal at this hypervariable position (figure 4a). Venoms from the sympatric neurotoxic snakes: Hemachatus haemachatus, Naja mossambica, and N. nivea were tested to ascertain if these substitutions reduce α-neurotoxin binding. Naja nubiae venom was tested to determine if there were any differences in binding patterns of sympatric and allopatric venoms. While Naja species are well described as feeding on other snakes [27], due to data deficient natural history observations, the degree to which H. haemachatus feeds upon snakes is unresolved, previous laboratory research has shown the venom had a greater affinity toward a snake (Coelognathus radiata) orthosteric mimotope than other taxa tested [22]. The native P. cana mimotope had significantly reduced susceptibility toward all the venoms tested (figure 4b). The 187W mutant was again tested to ensure the introduction of a serine (S) in place of the W typically present at position 187 did not affect binding, and that the resistance is indeed conferred by the presence of the positively charged 191K and 196K residues, (figure 4b). As with P. bivittatus (figure 3b), the substitution with the ancestral 191D significantly increased the binding across all venoms (figure 4b). Conversely, the substitution with the ancestral 196T increased binding marginally by N. nivea and N. mossambica, while the allopatric N. nubiae had a strong increase in binding (figure 4b). These data suggest that for sympatric venoms both the 191K and 196K play a role in providing the reduced susceptibility to the venoms albeit to varying degrees, with the 191K charge reversal providing a significantly greater resistance than 196K mutation which did not result in a charge reversal. The large difference in binding of the 196T analogue between N. nubiae and the other species venoms further supports the idea that proportions of specific 3FTx isoforms play a role in the overall target specificity of the crude venom [22]. Mole snakes are sympatric to many ophiophagus Naja, and P. cana was found in stomach contents of Naja anchietae [27]. Yet, little research has been conducted on the predation of mole snakes by Naja. There is a non-peer reviewed observation of N. nivea repeatedly biting P. cana with no noticeable envenomation effects and P. cana escaping seemingly unharmed after an hour of battling with the cobra [28]. Although this observation is unpublished, this natural history observation supports our laboratory finding that that P. cana has a reduced susceptibility toward Naja venom due to the coevolutionary arms race for resistance to evolve.
Figure 4.
The effects of venom from four elapid species against the Pseudaspis cana (native and mutants). (a) Amino acid sequences of native and mutant mimotopes. (b) Bar graphs represent the mean area under the curve (AUC) values of the adjacent curve graphs. Curve graphs show the mean wavelength (nm) shift in light with increased binding of venoms over a 120 s association phase. Each venom was tested in triplicate (n = 3). Error bars on all graphs represent the SEM. AUC values were statistically analysed using a one-way ANOVA with a Dunnett's multiple comparisons post hoc test comparing to the native mimotope. Statistical significance is indicated by an above the corresponding bar with a significance threshold of p < 0.01. All raw data and statistical analyses outputs can be found in electronic supplementary material, data S3. (Online version in colour.)
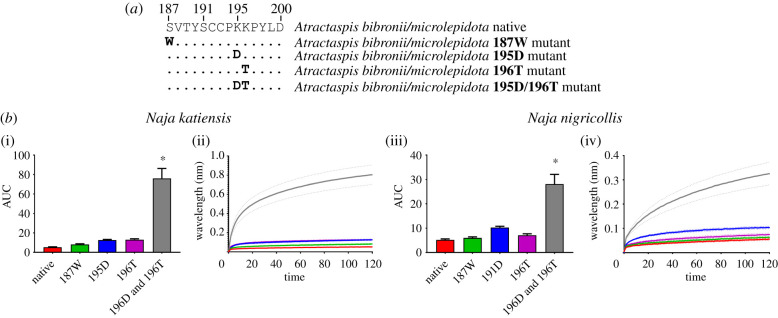
The last common ancestor of Atractaspis bibronii and A. microlepidota have evolved 195K and 196K at adjacent positions. It is only the 195K which is a charge reversal from their closest relative, with the positively charged K replacing the negatively charged D (figure 2). Again, the 187S mutant was tested to ensure this replacement of the ancestral 187 W played no role in reduced α-neurotoxin binding (figure 5b). Venoms from sympatric species, N. katiensis and N. nigricollis that are potential predators of A. bibronii/A. microlepidota were tested across the native and mutant mimotopes. There was no difference in binding from the native mimotopes with the 195K and 196K when each position individually replaced with the corresponding ancestral amino acid (figure 5b). However, once both positions were replaced by ancestral residues, a significant increase in the binding of both venoms occurred (figure 5b). This again suggests a synergistic effect of the multiple charged residues. Evolving Ks in adjacent positions might allow for resistance to be maintained if a random mutation removes one of the Ks at either position. It is uncertain if Naja spp. are predators of Atractaspis since natural history dietary data is deficient, however, since Naja are known to be non-specialist snake-eaters then such predation events are certainly possible. Future natural history field studies will be required to confirm this. Further to this, Atractaspis are also highly venomous [29–32], having been shown to contain 3FTxs [32] and thus these mutations may have evolved as a form of autoresistance to their own neurotoxins, as suggested for the N-glycosylation mode of autoresistance in elapids [5,6,15]. Similarly, Azemiops feae has the same KK doublet at positions 195 and 196 (figure 5) which in light of its documented neurotoxicity [21,33] is also suggestive of neurotoxicity autoresistance.
Figure 5.
The effects of venom from two sympatric Naja species against Atractaspis bibronii/A. microlepidota native and mutant mimotope sequences. (a) Amino acid sequences of native and mutant mimotopes. (b) Bar graphs represent the mean area under the curve (AUC) values of the adjacent curve graphs. Curve graphs show the mean wavelength (nm) shift in light with increased binding of venoms over a 120 s association phase. Each venom was tested in triplicate (n = 3). Error bars on all graphs represent the SEM. AUC values were statistically analysed using a one-way ANOVA with a Dunnett's multiple comparisons post hoc test comparing to the native mimotope. Statistical significance is indicated by an above the corresponding bar with a significance threshold of p < 0.01. All raw data and statistical analyses outputs can be found in electronic supplementary material, data S4. (Online version in colour.)
3. Conclusion
In summary, we have documented, for the first time, a novel form of resistance to snake venom neurotoxins that is driven by the replacement of ancestral negatively charged amino acids with the positively charged amino acid lysine (K) within the orthosteric site of the α-1 nicotinic acetylcholine receptor. The negatively charged α-1 orthosteric site provided the selection pressure for the evolution of snake venom neurotoxins with positively charged molecular surfaces that facilitate binding via opposite-charge interactions. Thus, mutations which result in a charge reversal, with the α-1 orthosteric site now having a net positive charge, electrostatically repel the positively charged neurotoxins. We have not only described this evolutionary novelty for the first time but suggested that this has occurred either as resistance by prey or as a mechanism of a snake being resistant to its own venom.
Conspicuously, our data also suggest that although the key positions confer reduced susceptibility individually, the majority of the resistance comes from a synergistic effect imposed by multiple positively charged positions at these key sites. Residue changes at these positions might also confer resistance through conformational shape change of the orthosteric site. If this conformational shape change does occur, future work should investigate how this affects the binding of acetylcholine since evolutionary trade-offs usually come at a fitness disadvantage and are selected against in the absence of a correspondingly greater benefit [34,35], as has been suggested for the N-glycosylated form of resistance described previously [15].
The resistance to both sympatric and allopatric α-neurotoxic snake venoms suggests that although the coevolutionary selection pressures are likely due to predator–prey interactions between these species, the reduced susceptibility is also present across the allopatric α-neurotoxin types (figures 1 and 3–5), signifying similar targeting mechanisms of these toxins, reinforcing the paradigm that sensitivities in the target provides the selection pressure for the evolution of the toxin.
Any discussion regarding the functional and evolutionary aspect of venom must be viewed in conjunction with the ecology of the venomous species and its natural predator/prey. This would give the most prospective interpretation of the results since in the majority of instances antagonistic relationships between predator and prey are what drive the evolution of such traits. This study documenting novel forms of resistance to snake venom neurotoxins provides an excellent foundation upon which to build more expansive evolutionary theories. It is clear that there is a huge gap in the literature regarding the predatory ecology of many of the species investigated in this study, and further work should include natural history observations to fill these knowledge gaps. Future research should sequence additional species across the animal kingdom in order to document how prevalent such charge reversal mutations are, beyond the 10 convergent evolutionary events, and if these correspond with other predator and prey coevolutionary arms races involving species that use α-neurotoxins.
4. Materials and methods
(a). Venom collection and preparation
All venom work was undertaken under University of Queensland Biosafety Approval #IBC134BSBS2015. Venoms were sourced from the pre-existing, long-term cryogenic collection of the Toxin Evolution Lab. All venom samples were lyophilized and reconstituted in double deionized water (ddH2O), and centrifuged (4°C, 10 min at 14 000 relative centrifugal force (RCF)). The supernatant was made into a working stock (1 mg ml−1) in 50% of glycerol at −20°C. The concentrations of experimental stocks were determined using a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Fisher, Sydney, Australia) at an absorbance wavelength of 280 nm.
(b). Mimotope production and preparation
Following methods from a previously developed assay [22,23], a 13–14 amino acid mimotope of the vertebrate α-1 nAChR orthosteric site was developed by GenicBio Ltd. (Shanghai, China) designed upon specification. Mimotopes from Python bivittatus, Pseudaspis cana and Atractaspis bibronii/A. microlepidotus were based on already published sequences[15]. The mimotope for Mellivora capensis was obtained from UniProt public database (accession A0A0P0D4F3).
The C-C of the native mimotope is replaced during peptide synthesis with S-S to avoid uncontrolled postsynthetic thiol oxidation. The C-C bond in the nAChR binding region does not participate directly in analyte-ligand binding [12,36,37], thus replacement to S-S is not expected to have any effect on the analyte-ligand complex formation. However, the presence of the C-C bridge is key in the conformation of the interaction site of whole receptors [38]. As such, we suggest direct comparisons of kinetics data, such as Ka or KD, between nAChR mimotopes and whole receptor testing should be avoided, or at least approached with caution. Mimotopes were further connected to a biotin linker bound to two aminohexanoic acid (Ahx) spacers, forming a 30 Å linker.
Mimotope dried stocks were solubilized in 100% dimethyl sulfoxide (DMSO) and diluted in ddH2O at 1 : 10 dilution to obtain a stock concentration of 50 µg ml−1. Stocks were stored at −80°C until required.
(c). Biolayer interferometry
BLI is a label-free, microfluidics-free, optical technique that precisely measures the thickness of biomolecules accumulating on the interaction surface of an optical fibre-coated biosensor. The binding of molecules to the biosensor causes a measurable spectral shift in the wavelength of light being reflected through the fibre optic biosensor, which yields quantitative, kinetic interaction information.
Full details of the developed assay, including all methodology and data analysis, can be found in the validated protocol [23] and further data using this protocol [21,22]. In brief, the BLI assay was performed on the Octet HTX system (ForteBio, Fremont, CA, USA). Venom samples were diluted 1 : 20, making a final concentration of 50 µg ml−1 per well. Mimotope aliquots were diluted 1 : 50, with a final concentration of 1 µg ml−1 per well. The assay running buffer was 1X DPBS with 0.1% BSA and 0.05% Tween-20. Preceding experimentation, Streptavidin biosensors were hydrated in the running buffer for 30–60 min, while on a shaker at 2.0 revolutions per minute (RPM). The dissociation of analytes occurred using a standard acidic glycine buffer solution (10 mM glycine (pH 1.5–1.7) in ddH2O). Raw data are provided in electronic supplementary material, data S1–S4.
(d). Data processing and analysis
All data obtained from BLI on Octet HTX system (ForteBio) were processed in exact accordance to the validation of this assay [23]. The association step data (a recording of the wavelength shift (nm) at 0.2 second intervals over a 120 s period) were obtained and imported into Prism 8.0 software (GraphPad Software Inc., La Jolla, CA, USA) where area under the curve (AUC) and one-way ANOVA with a Dunnett's multiple comparisons analyses were conducted and graphs produced. Statistical analysis data is available in electronic supplementary material, data S1–S4.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Ethics
No live animals were worked with in this research; therefore, ethics approval was not required.
Data accessibility
Raw data have been included as electronic supplementary material.
Authors' contributions
R.J.H. and B.G.F. designed and conceptualized the research. R.J.H. performed the experiments and analysed the data. B.G.F. supervised the research and provided resources. R.J.H. wrote the manuscript. R.J.H. and B.G.F. further edited subsequent drafts of the manuscript.
Competing interests
The authors declare no competing interests.
Funding
This work was supported by Australian Research Council Discovery Project DP210102406 for B.G.F. and the University of Queensland International PhD scholarship fund for R.J.H.
References
- 1.Dawkins R, Krebs JR. 1979. Arms races between and within species. Proc. R. Soc. Lond. B: Biol. Sci. 205, 489–511. ( 10.1098/rspb.1979.0081) [DOI] [PubMed] [Google Scholar]
- 2.Van Valen L 1973. A new evolutionary law. Evol. Theory 1, 1–30. [Google Scholar]
- 3.Arbuckle K, de la Vega RCR, Casewell NR. 2017. Coevolution takes the sting out of it: evolutionary biology and mechanisms of toxin resistance in animals. Toxicon 140, 118–131. ( 10.1016/j.toxicon.2017.10.026) [DOI] [PubMed] [Google Scholar]
- 4.Drabeck DH, Dean AM, Jansa SA. 2015. Why the honey badger don't care: convergent evolution of venom-targeted nicotinic acetylcholine receptors in mammals that survive venomous snake bites. Toxicon 99, 68–72. ( 10.1016/j.toxicon.2015.03.007) [DOI] [PubMed] [Google Scholar]
- 5.Takacs Z, Wilhelmsen KC, Sorota S. 2001. Snake α-neurotoxin binding site on the Egyptian cobra (Naja haje) nicotinic acetylcholine receptor is conserved. Mol. Biol. Evol. 18, 1800–1809. ( 10.1093/oxfordjournals.molbev.a003967) [DOI] [PubMed] [Google Scholar]
- 6.Takacs Z, Wilhelmsen KC, Sorota S. 2004. Cobra (Naja spp.) nicotinic acetylcholine receptor exhibits resistance to erabu sea snake (Laticauda semifasciata) short-chain α-neurotoxin. J. Mol. Evol. 58, 516–526. ( 10.1007/s00239-003-2573-8) [DOI] [PubMed] [Google Scholar]
- 7.Barchan D, Kachalsky S, Neumann D, Vogel Z, Ovadia M, Kochva E, Fuchs S. 1992. How the mongoose can fight the snake: the binding site of the mongoose acetylcholine receptor. Proc. Natl Acad. Sci. USA 89, 7717–7721. ( 10.1073/pnas.89.16.7717) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barchan D, Ovadia M, Kochva E, Fuchs S. 1995. The binding site of the nicotinic acetylcholine receptor in animal species resistant to alpha-bungarotoxin. Biochemistry 34, 9172–9176. ( 10.1021/bi00028a029) [DOI] [PubMed] [Google Scholar]
- 9.Kachalsky SG, Jensen BS, Barchan D, Fuchs S. 1995. Two subsites in the binding domain of the acetylcholine receptor: an aromatic subsite and a proline subsite. Proc. Natl Acad. Sci. USA 92, 10 801–10 805. ( 10.1073/pnas.92.23.10801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bracci L, Lozzi L, Lelli B, Pini A, Neri P. 2001. Mimotopes of the nicotinic receptor binding site selected by a combinatorial peptide library. Biochemistry 40, 6611–6619. ( 10.1021/bi0023201) [DOI] [PubMed] [Google Scholar]
- 11.Dellisanti C, Yao Y, Stroud JC, Wang Z-Z, Chen L. 2007. Structural determinants for α-neurotoxin sensitivity in muscle nAChR and their implications for the gating mechanism. Channels 1, 234–237. ( 10.4161/chan.4909) [DOI] [PubMed] [Google Scholar]
- 12.Tzartos S, Remoundos MS. 1990. Fine localization of the major alpha-bungarotoxin binding site to residues alpha 189–195 of the Torpedo acetylcholine receptor: residues 189, 190, and 195 are indispensable for binding. J. Biol. Chem. 265, 21 462–21 467. [PubMed] [Google Scholar]
- 13.Dellisanti CD, Yao Y, Stroud JC, Wang Z-Z, Chen L. 2007. Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94 Å resolution. Nat. Neurosci. 10, 953–962. ( 10.1038/nn1942) [DOI] [PubMed] [Google Scholar]
- 14.Suryamohan K, et al. 2020. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 52, 106–117. ( 10.1038/s41588-019-0559-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan MA, et al. 2020. Widespread evolution of molecular resistance to snake venom α-neurotoxins in vertebrates. Toxins 12, 638 ( 10.3390/toxins12100638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kini RM, Doley R. 2010. Structure, function and evolution of three-finger toxins: mini proteins with multiple targets. Toxicon 56, 855–867. ( 10.1016/j.toxicon.2010.07.010) [DOI] [PubMed] [Google Scholar]
- 17.Rahman MM, Teng J, Worrell BT, Noviello CM, Lee M, Karlin A, Stowell MH, Hibbs RE. 2020. Structure of the native muscle-type nicotinic receptor and inhibition by snake venom toxins. Neuron 106, 952–962.e5. ( 10.1016/j.neuron.2020.03.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Begg C, Begg K, Du Toit J, Mills M. 2003. Sexual and seasonal variation in the diet and foraging behaviour of a sexually dimorphic carnivore, the honey badger (Mellivora capensis). J. Zool. 260, 301–316. ( 10.1017/S0952836903003789) [DOI] [Google Scholar]
- 19.Barber CM, Isbister GK, Hodgson WC. 2013. Alpha neurotoxins. Toxicon 66, 47–58. ( 10.1016/j.toxicon.2013.01.019) [DOI] [PubMed] [Google Scholar]
- 20.Utkin Y, Sunagar K, Jackson T, Reeks T, Fry B. 2015. Three-finger toxins (3FTxs). In Venomous reptiles and their toxins: evolution, pathophysiology and biodiscovery (ed. Fry B), pp. 215–227. New York, NY: Oxford University Press. [Google Scholar]
- 21.Harris RJ, Zdenek CN, Debono J, Harrich D, Fry BG. 2020. Evolutionary interpretations of nicotinic acetylcholine receptor targeting venom effects by a clade of Asian Viperidae snakes. Neurotox. Res. 38, 312–318. ( 10.1007/s12640-020-00211-2) [DOI] [PubMed] [Google Scholar]
- 22.Harris RJ, Zdenek CN, Harrich D, Frank N, Fry BG. 2020. An appetite for destruction: detecting prey-selective binding of α-neurotoxins in the venom of Afro-Asian elapids. Toxins 12, 205 ( 10.3390/toxins12030205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zdenek CN, et al. 2019. A taxon-specific and high-throughput method for measuring ligand binding to nicotinic acetylcholine receptors. Toxins 11, 600 ( 10.3390/toxins11100600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Shea M 2018. The book of snakes: a life-size guide to six hundred species from around the world. Chicago, IL: University of Chicago Press. [Google Scholar]
- 25.Wilson SK, Swan G. 2017. A complete guide to reptiles of Australia, 5th edn Sydney, Australia: New Holland Publishers. [Google Scholar]
- 26.Ehmann H 1993. Boidae. In Fauna of Australia, pp. 1–15. Clayton, Australia: CSIRO Publishing. [Google Scholar]
- 27.Shine R, Branch W, Webb J, Harlow PS, Shine T, Keogh JS. 2007. Ecology of cobras from southern Africa. J. Zool. 272, 183–193. ( 10.1111/j.1469-7998.2006.00252.x) [DOI] [Google Scholar]
- 28.Mitchell G, Mitchell C. 2019. Remarkable sighting: cape cobra versus mole snake. Africa Geographic; See https://africageographic.com/stories/remarkable-sighting-cape-cobra-versus-mole-snake. [Google Scholar]
- 29.Oulion B, et al. 2018. Factor X activating Atractaspis snake venoms and the relative coagulotoxicity neutralising efficacy of African antivenoms. Toxicol. Lett. 288, 119–128. ( 10.1016/j.toxlet.2018.02.020) [DOI] [PubMed] [Google Scholar]
- 30.Tilbury CR, Verster J. 2016. A fatal bite from the burrowing asp Atractaspis corpulenta (Hallowell 1854). Toxicon 118, 21–26. ( 10.1016/j.toxicon.2016.04.035) [DOI] [PubMed] [Google Scholar]
- 31.Weiser E, Wollberg Z, Kochva E, Lee S. 1984. Cardiotoxic effects of the venom of the burrowing asp, Atractaspis engaddensis (Atractaspididae, Ophidia). Toxicon 22, 767–774. ( 10.1016/0041-0101(84)90159-4) [DOI] [PubMed] [Google Scholar]
- 32.Terrat Y, et al. 2013. Atractaspis aterrima toxins: the first insight into the molecular evolution of venom in side-stabbers. Toxins 5, 1948–1964. ( 10.3390/toxins5111948) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Utkin YN, et al. 2012. Azemiopsin from Azemiops feae viper venom, a novel polypeptide ligand of nicotinic acetylcholine receptor. J. Biol. Chem. 287, 27 079–27 086. ( 10.1074/jbc.M112.363051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brodie ED III, Brodie ED Jr. 1999. Costs of exploiting poisonous prey: evolutionary trade-offs in a predator-prey arms race. Evolution 53, 626–631. ( 10.1111/j.1558-5646.1999.tb03798.x) [DOI] [PubMed] [Google Scholar]
- 35.Blanchard BD, Moreau CS. 2017. Defensive traits exhibit an evolutionary trade-off and drive diversification in ants. Evolution 71, 315–328. ( 10.1111/evo.13117) [DOI] [PubMed] [Google Scholar]
- 36.McLane KE, Wu X, Conti-Tronconi BM. 1994. An α-bungarotoxin-binding sequence on the Torpedo nicotinic acetylcholine receptor α-subunit: Conservative amino acid substitutions reveal side-chain specific interactions. Biochemistry 33, 2576–2585. ( 10.1021/bi00175a029) [DOI] [PubMed] [Google Scholar]
- 37.McLane KE, Wu X, Diethelm B, Conti-Tronconi BM. 1991. Structural determinants of α-bungarotoxin binding to the sequence segment 181–200 of the muscle nicotinic acetylcholine receptor. α-subunit: effects of cysteine/cystine modification and species-specific amino acid substitutions. Biochemistry 30, 4925–4934. ( 10.1021/bi00234a013) [DOI] [PubMed] [Google Scholar]
- 38.Testai FD, Venera GD, Peña C, de Jiménez Bonino MJB. 2000. Histidine 186 of the nicotinic acetylcholine receptor α subunit requires the presence of the 192–193 disulfide bridge to interact with α-bungarotoxin. Neurochem. Int. 36, 27–33. ( 10.1016/S0197-0186(99)00099-6) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data have been included as electronic supplementary material.