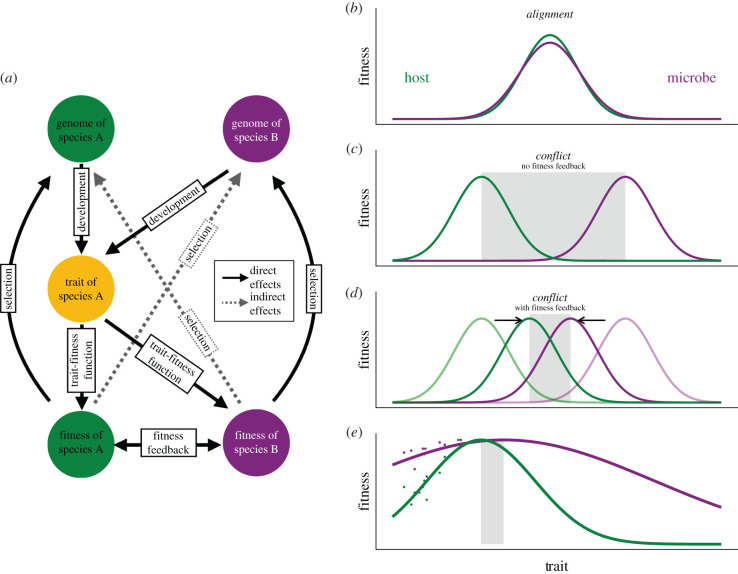
Figure 1.
(a) The genomes of two species, A (host, green) and B (microbe, purple), affect a joint trait (yellow) that can affect the fitness of both. Fitness of A and B results in direct selection on the genomes of A and B, respectively. (b) The relationship between the trait and fitnesses of hosts and microbes when both experience stabilizing selection for the same trait value. This aligns fitness interests regardless of expressed trait values in the population. (c) A difference between species A and B in the optimal value of the trait. This results in conflict across the range of trait values between the two optima (grey shading). (d) Such conflicts can be ameliorated or resolved by indirect effects of traits on each species’ fitness via positive fitness feedbacks (see text), which effectively move the trait optima together (indicated by arrows). (e) Data (points) from Haney et al. [28] for host (Arabidopsis thaliana) and microbe (Pseudomonas fluorescens) fitness as a function of a joint trait, plant root branching. Lines show data fit to a Gaussian curve for each species, which predict different optima, potentially creating conflict (grey-shaded region) and stronger direct selection on plants (curve steepness).