Figure 2.
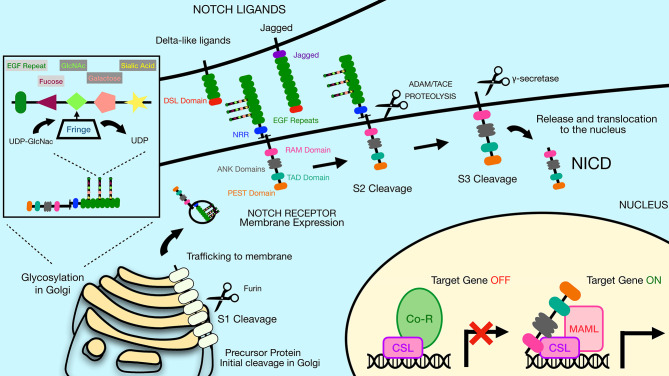
The Notch signaling pathway is mediated by a series of proteolytic events. Notch receptors are generated by ribosomes bound to the endoplasmic reticulum (ER) and trafficked through the Golgi to the plasma membrane. During the time in the Golgi, Notch receptors are cleaved at Site 1 (S1) by furin-like proteases to generate two sub-parts that remain non-covalently associated. The N-terminal subunit can be O-glycosylated via the activity of a series of glycosyltransferase enzymes, including members of the Fringe family that catalyze addition of N-acetylglucosamine residues residues to the glycan chain. Once at the plasma membrane, Notch receptors are inactive unless bound by ligand on adjacent cells. Notch ligands constitute two families, the Delta-like ligands and the Jagged family ligands. Both types of ligands contain a conserved Delta-Serrate ligand (DSL) domain that mediates binding to Notch receptors. Upon ligand binding, Notch receptors undergo cleavage by ADAM family proteins at Site 2 (S2). This allows subsequent cleavage by γ-secretase at Site 3 (S3), releasing the Notch intracellular domain (NICD). The liberated NICD is translocated into the nucleus to bind to RBP-Jκ along with the coactivator Maml, leading to activation of target gene expression. Prior to binding of NICD and Maml, RBP-Jκ is associated with co-repressor proteins that prevent transcription of target genes. DSL, Delta/Serrate/Lag2; UDP, Uridine diphosphate; GlcNAc, N-acetylglucosamine; NICD, Notch intracellular domain; Maml, mastermind-like protein; Co-R, corepressor protein complexes.