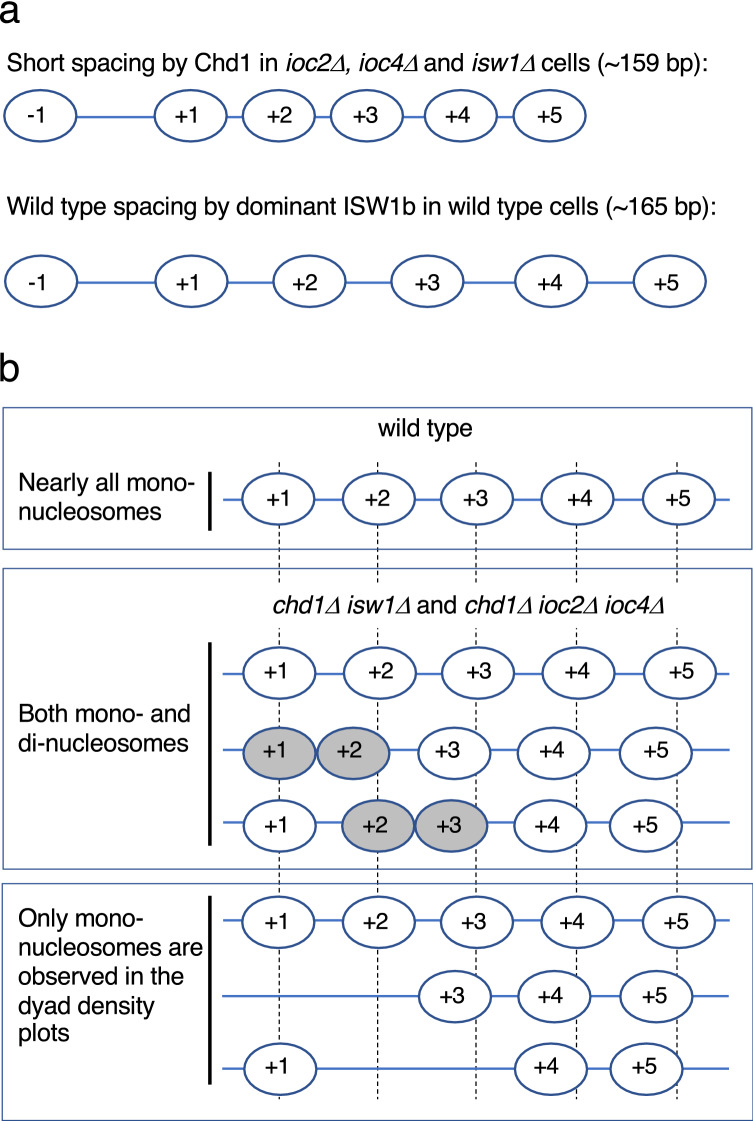
Figure 6.
Dominant role of ISW1b in chromatin organisation. (a) Global average nucleosome spacing is primarily determined by ISW1b (wild type cells), since loss of ISW1a has little effect. Short spacing is attributed to Chd1, because the chd1∆ isw1∆ double mutant has highly disrupted chromatin. (b) Dinucleosomes disrupt phasing and reduce spacing. Model to show the effects of dinucleosomes on chromatin organisation. Top panel: Average nucleosome positions on a gene in wild type cells with regular spacing. Middle: Close-packed dinucleosomes (grey ovals) are high in the absence of both ISW1b and Chd1. These dinucleosomes preferentially involve the + 2 nucleosome i.e., they are mostly + 1/+ 2 or + 2/+ 3 dinucleosomes. Nucleosomes farther down the gene (+ 3 etc.) may be regularly spaced relative to the dinucleosome, but because a linker is missing in the dinucleosome, downstream nucleosomes are out of phase with other nucleosomal arrays, resulting in poor phasing due to interference patterns from the different arrays. Bottom: Dinucleosomes are absent from the mononucleosome dyad density plots, resulting in depressed, flattened peaks. These effects are strongest in cells lacking both ISW1b and Chd1. Dinucleosomes that cannot be resolved by ISW1b can account for shorter spacing in chd1∆ cells.