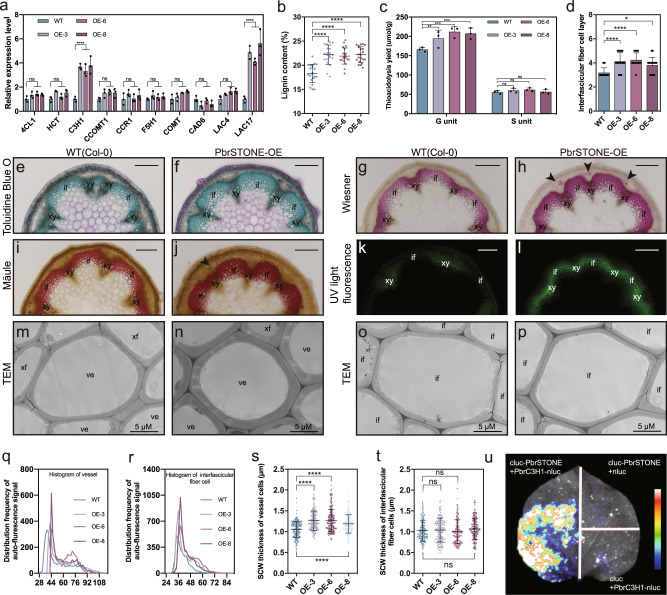
Fig. 6. Functional validation of PbrSTONE in Arabidopsis.
a Expression levels of genes encoding enzymes involved in monolignol biosynthesis and polymerization in WT and PbrSTONE transgenic Arabidopsis plants using qPCR analysis; n = 3 biologically independent samples. b, c Lignin analysis for WT and PbrSTONE transgenic Arabidopsis plants; h: n = 21 biologically independent samples for b, and n = 3 for c. d Layers of stem interfascicular fiber cells in transgenic Arabidopsis plants expressing PbrSTONE; n = 16 biologically independent samples. e, f Cross-sections of 8-week-old inflorescence stems stained with Toluidine blue O. g–j Cross-sections of 8-week-old inflorescence stems stained with Mäule and Wiesner reagent, which bind to S-lignin monomer and G-lignin monomer, respectively. k, l Lignin auto-fluorescence of cross-sections detected under UV light. Scale bars: 200 μm in k–r. Bar in k–r = 200 μm; xy, xylem; if, interfascicular fiber cell. The transgenic and WT plants were cultivated two times and cross-sections were taken from five independent plants which showed similar results. A representative picture from each line is shown. m–p Transmission electron micrographs of the cross-section of cells in e; ve, vessel; xf, xylary fiber; if, interfascicular fiber cells. q, r Statistical analysis of lignin auto-fluorescence signal in the vessel cells and interfascicular fiber cells. More than 12 samples in each line were analyzed. s, t Statistical analysis of secondary cell wall thickness in the vessel cells and interfascicular fiber cells. More than 50 cells in each line were analyzed. u Firefly luciferase complementation assay in young Nicotiana benthamiana leaves. Data presented are mean ± SD; p-values were determined by two-tailed Student’s t-test (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; ns, not significant). Source data underlying (a–t) are provided as a Source Data file.