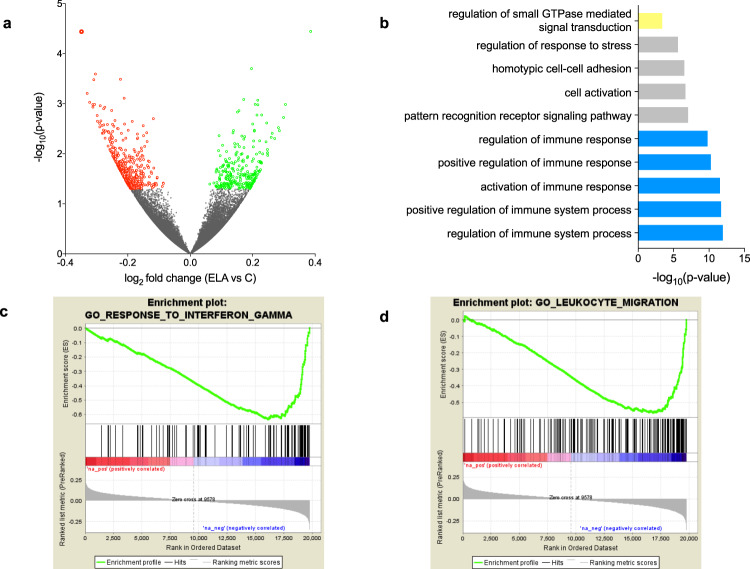
Fig. 7. Differential gene expression in subjects with a history of early-life adversity (ELA).
a Volcano plot of RNA-Seq data showing the 261 and 474 genes that were up- (green circles) or downregulated (red circles) in the ELA group compared with the control (C) group (GLM model using gender, age, pH, PMI, and RIN as covariates; nominal P value < 0.05). b Gene Ontology analysis of the 735 differentially expressed genes in the ELA group. Terms showing evidence of enrichment for differential methylation, histone profile or chromatin state are shown in yellow (small GTPase) or blue (immune processes; see also Supplementary Fig. 16f). c, d Gene Set Enrichment Analysis (GSEA) of gene expression changes in ELA subjects. Genes were ranked based on log2 fold changes from the C versus ELA differential expression analysis (“Ranked list metric”, in grey in the lower portion of each panel). Genes with the highest positive fold changes (in red, upregulated in the ELA group) are at the extreme left of the distribution, and those with the lowest negative fold changes (in blue, downregulated in the ELA group) are at the extreme right. A running enrichment score (green line, the upper portion of each panel) was computed for gene sets from the MSigDB curated molecular signatures database and used to identify enriched gene sets42. Among the numerous gene sets related to the immune function that showed evidence of genome-wide significant negative correlation with ELA (see main text, and Supplementary Table 14), two representative gene sets are shown (with the middle portion of each panel showing vertical black lines where members of the gene set appear in the ranked list of genes): c “Interferon-gamma”, and d “Leukocyte migration”. Of note, an oligodendrocyte-specific gene collection, which we recently found downregulated in the anterior cingulate cortex of subjects with a history of ELA40, positively correlated with ELA in the amygdala (see Supplementary Fig. 16d), suggesting opposite adaptations in this glial population between cortical and subcortical structures. Source data are provided as a Source Data file.