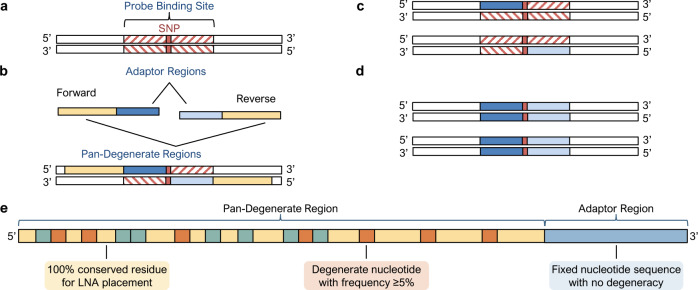
Fig. 1. Overview of PANDAA method and primer design.
a Heterogeneous genomes contain single-nucleotide variants within the probe-binding site that prevent the use of probe-based qPCR for DRM discrimination. b PANDAA primers overlap with the probe-binding site, adapting the nucleotides proximal to the DRM of interest that would otherwise abrogate probe hybridization. c, d As qPCR proceeds, the newly generated amplicon will contain probe-binding sites flanking the DRM that are perfectly complementary to the probe. e PANDAA primers contain two key features: a 3′ ADR that is matched to the probe-binding site and a pan-degenerate (PDR) region that incorporates the nucleotide degeneracy observed in the primer-binding site of the target. The PDR accounts for the high degree of variability in primer-binding sites. LNA bases at 100% conserved positions in the PDR offset the thermodynamic instability of mismatches between the primer ADR and the template.