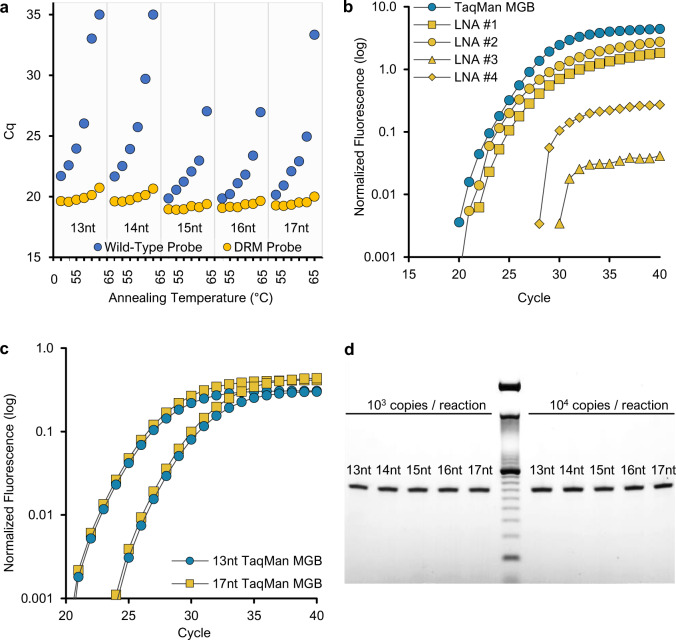
Fig. 2. Optimization of PANDAA probe design.
a 13–17-nt TaqMan-MGB probes for 103 wild-type and DRM codons. Sensitivity and specificity were evaluated using a 55–65 °C annealing temperature gradient. Although predicted Tm values ranged from 59 to 63 °C, there was little impact sensitivity with the AAC probes (yellow circles) for any probe length as the annealing temperature increased; however, non-specific hybridization of the AAA probe (blue circles) to the mismatched AAC template was reduced as annealing temperature increased. b The performance of four 5′ hydrolysis probes with various placement of LNA nucleotides (yellow) was compared to a TaqMan-MGB probe (blue) of the same length. LNAs were found to have comparable performance to MGB probes although LNA nucleotide placement also had to be determined empirically. c Increasing probe length did not reduce sensitivity, as 13-nt probes (blue circles) had similar Cq values to 17-nt probes (yellow) at 104 and 103 copies per reaction. d To confirm that amplification efficiency was not proportionally inhibited as the number of overlapping nucleotides increased, PANDAA reactions utilizing 13–17-nt probes were resolved on a 4% agarose gel, demonstrating comparable band intensities of a 66-bp amplicon across all probe lengths at both 103 and 104 DNA copies per reaction (middle lane: 10-bp DNA ladder). Furthermore, no non-specific products were evident.