Fig. 3. Optimization of PANDAA primer design.
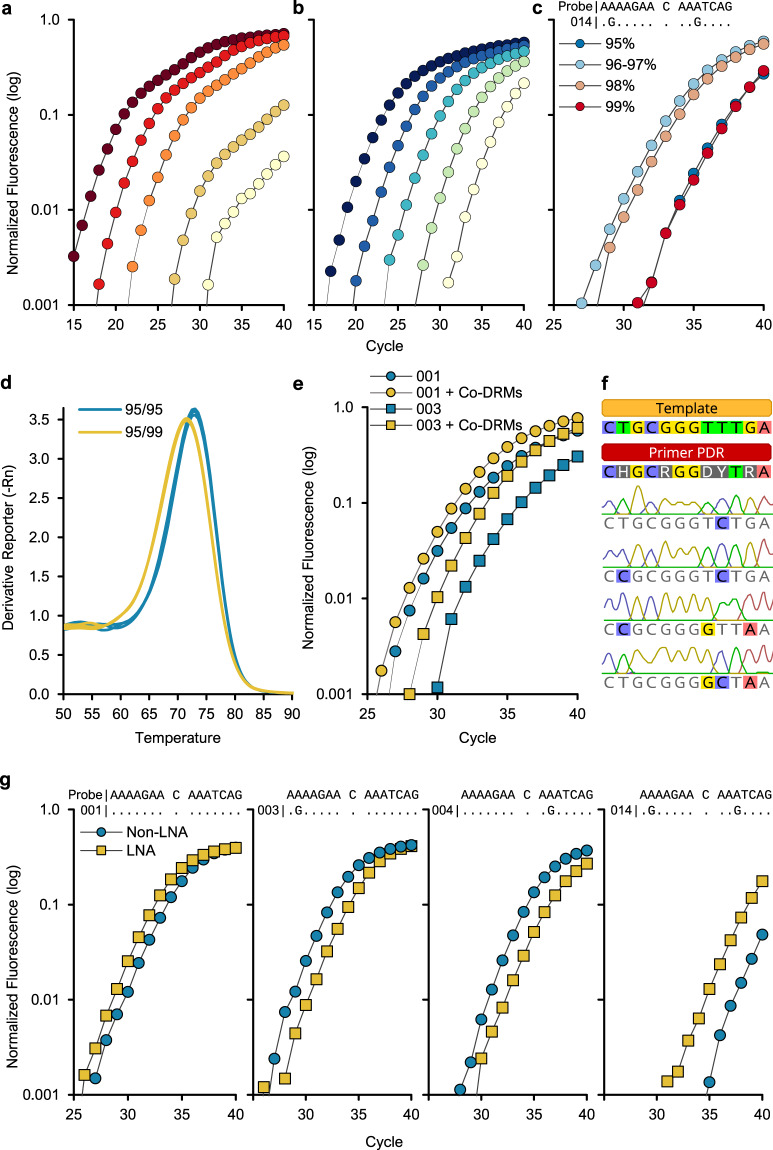
Consensus versus PANDAA primers. DNA template 001 was diluted from 106 to 100 copies per reaction and was amplified using either the a majority consensus primers with no degenerate bases or b PANDAA primers. Both sets of primers contained the same 3′ ADR. A single mismatch was present in both the forward and reverse consensus primers (Supplementary Table 4). Using the K103N DRM-specific probe, both primer sets showed similar sensitivity down to 104 copies; however, the lowest copy number quantified using consensus primers was 104 copies compared to 100 copies for the PANDAA primers. Although <104 copies were detectable using consensus primers, the copies were not accurately quantifiable. PDR degeneracy, co-expressed DRMs, and melt curve: c Using 103 copies of template 014, which contains a mismatch in each ADR, optimal PDR degeneracy was determined empirically. d The 2830F-99% PDR variant with either the 2896R-95% (blue) or 2896R-99% (yellow) variant in a SYBR green qPCR with the 014 template showed a similar relationship between PDR degeneracy and reaction efficiency to that with probe-based PANDAA. The lower amplicon Tm and broader melt curve with the 2896R-99% primer reflected the wider range of predicted GC% content compared to 2896R-95% (25.8–58.1% versus 29.0–54.8%, respectively), and therefore the wider primer Tm range (63.0–78.7 °C versus 64.2–77.3 °C, respectively). e Additional degeneracy was introduced at co-expressed DRMs included codons 100 and 101 in primer 2830 F and 106 in 2896 R. Performance was evaluated using template 001 (circles) or template 003 (squares). Primers with co-expressed DRMs (yellow) compared to the previous iteration of PANDAA primers (blue) were found to improve sensitivity. f Single-clone sequencing after PANDAA on a homogeneous DNA template demonstrated that adaptation was occurring within the PDR at positions containing degenerate bases. g Inclusion of LNA nucleotides at 100% conserved positions in the PDR increased sensitivity in templates containing probe-binding site mismatches in the forward ADR, reverse ADR, or both.