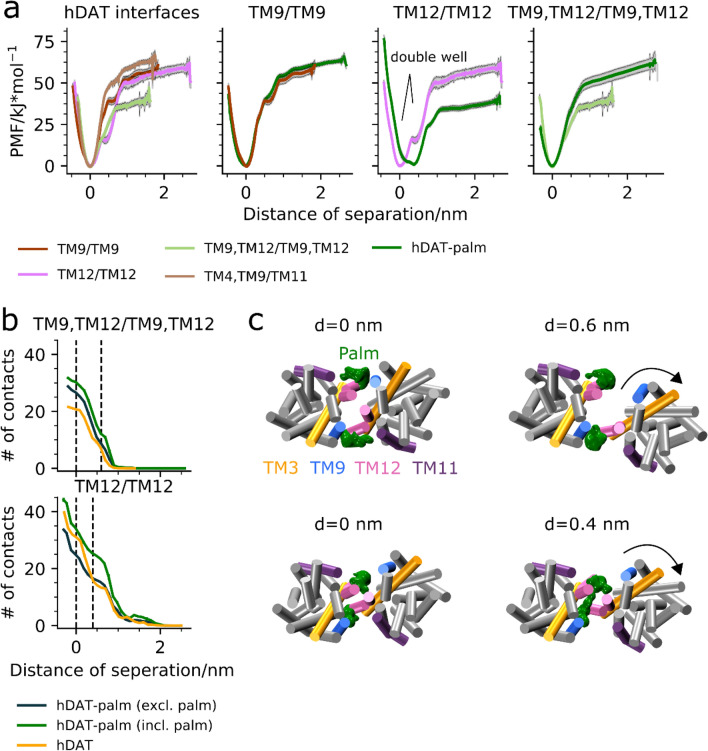
Figure 3.
Potential of mean force energy profiles and contact analysis of hDAT and hDAT-palm interfaces. Illustrated in the first panel in (a) are all the PMFs of the different hDAT interfaces: TM9/TM9 (brown), TM9,TM12/TM9,TM12 (light green), TM12,TM12 (pink), and TM4, TM9/TM11 (light brown). Shown in the subsequent plots are PMF comparisons of the interfaces TM9/TM9, TM9,TM12/TM9,TM12, and TM12/TM12 between hDAT and hDAT-palm (green). Shown in (b) are the average number of residue contacts calculated within 1 Å distance bins along hDAT dimer separation relative to the distance at energy minimum as detected from the PMF energy profiles. The number of residue contacts for the two systems, hDAT and hDAT-palm, at the two interfaces, TM12/TM12 and TM9, TM12/TM9, TM12, were monitored. For both interfaces the Cys581 residue where palm is attached is either included (incl.) or excluded (excl.) from the contacts analysis. A contact was counted when the distance between two residue pairs was below 7 Å. Each contact was unbiased. Error bars have been included by performing bootstrapping on the data using 10 iterations, although they are barely visible. Shown in (c) are representative dimer conformations of hDAT-palm at different separation distances for the TM9, TM12/TM9, TM12 interface (top) and the TM12/TM12 interface (bottom). Highlighted are helices TM3, TM9, TM11 and TM12. The green mesh corresponds to the palm occupancies calculated using the Volmap plugin in VMD for the dimer conformations found around the given separation distance. For the Volmap calculations the size of the system beads were set to 2.6 Å and the “occupancy” setting was selected. The arrows indicate the movement of the proteins with respect to each other.