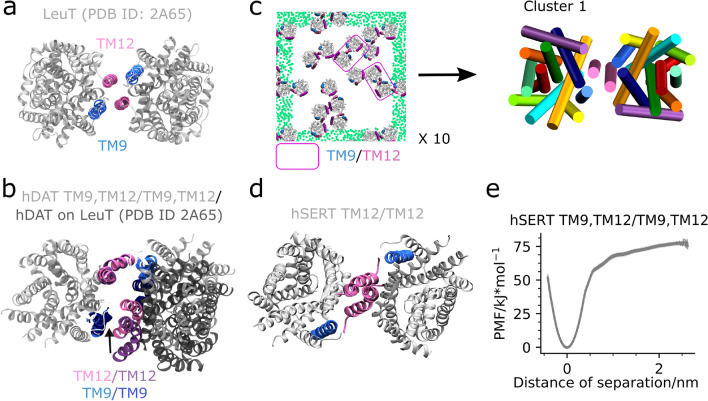
Figure 6.
LeuT and MATs have a similar TM12/TM12 and TM12,TM9/TM12,TM9 interface. Shown in (a) is a top view of the LeuT crystal structure dimer (PDB ID: 2A65) with helices TM9 and TM12 highlighted. In (b) the central hDAT dimer structure from cluster 1 (see Fig. 2b) is superimposed on hDAT monomers aligned to the LeuT dimer. Depicted in (c) is a representative last frame of the 10 LeuT self-assembly repeat simulations (MD1). See Supplementary Fig. S18 for a full overview. Each system contains 16 LeuT (PDB ID: 2A65) proteins equally spaced but randomly orientated in a pure POPE membrane and has been simulated for 30 µs. The helices TM9, TM11, and TM12 are highlighted in blue, purple, and mauve, respectively. Emphasized by pink boxes are dimers containing TM12/TM12 and to some extent TM9 contacts. The green dots correspond to the GL1 bead in POPE and are shown for the nearest periodic image to the system. For the central simulation box the dots have been omitted. To the right in (c) is a top view with respect to the membrane normal of the central dimer conformation observed in cluster 1. Notice that the dimer achieved from our simulations has an interface consisting of TM12/TM12, TM9 contacts, due to a slight change in the two proteins relative orientation. This change in orientation relative to the LeuT crystal structure dimer results in a loss of contacts to one of the TM9 helices. Represented in (d) is the stable hSERT TM12/TM12 dimer previously detected14. Finally, in (e) is the PMF of the hSERT TM9,TM12/TM9,TM12 interface.