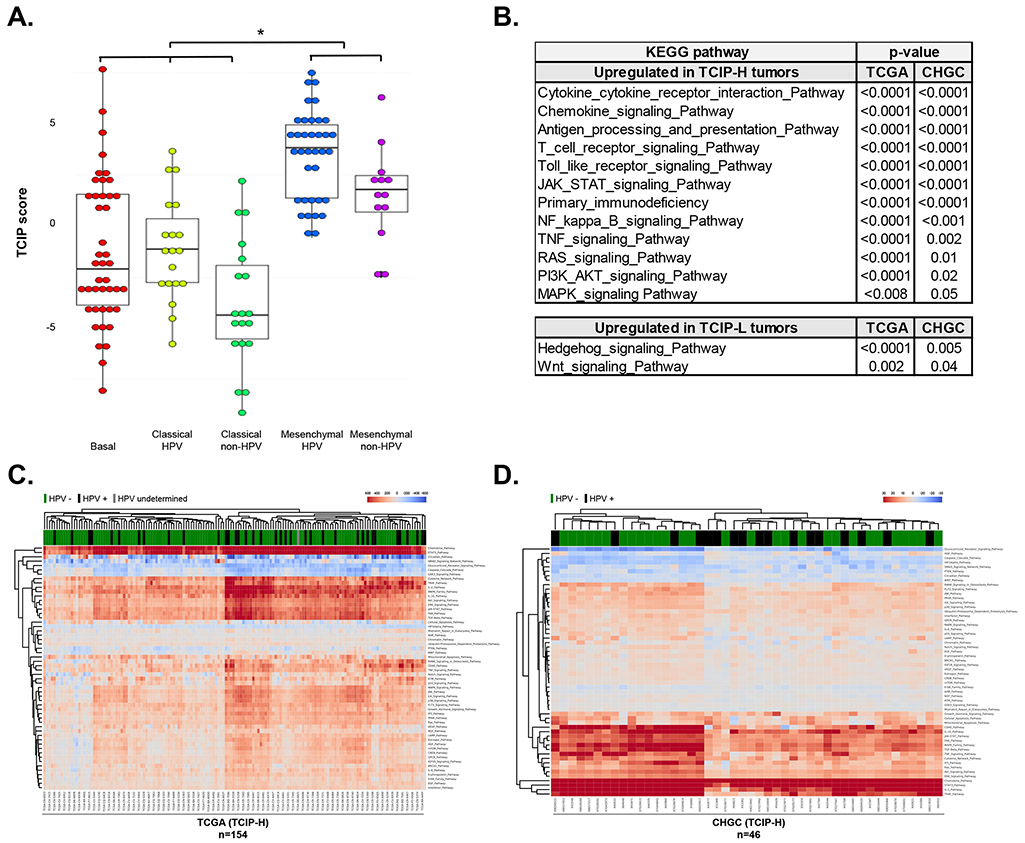
Figure 5. Correlation of T-cell inflamed phenotypes with SCCHN expression subtypes and signaling pathways.
(A) TCIP-H tumors correlated significantly with the mesenchymal subtype, while the TCIP-L tumors correlated with the basal and classical subtypes, irrespective of HPV status (CHGC). (B) Table summarizing the results of gene set enrichment analysis for pathways upregulated in TCIP-H and TCIP-L SCCHN tumors. (C-D) RNA-Seq data (for TCGA cohort, C) or gene expression microarray data (for CHGC cohort, D) was processed and analyzed using the iPANDA software suite. The hierarchically clustered heatmap depicts main signaling pathways dysregulated in TCIP-H samples (transcriptomic data derived from TCIP-L tumors was used as a reference for analysis). Downregulated iPANDA values for each sample/pathway are indicated in blue, while upregulated values are shaded in red.