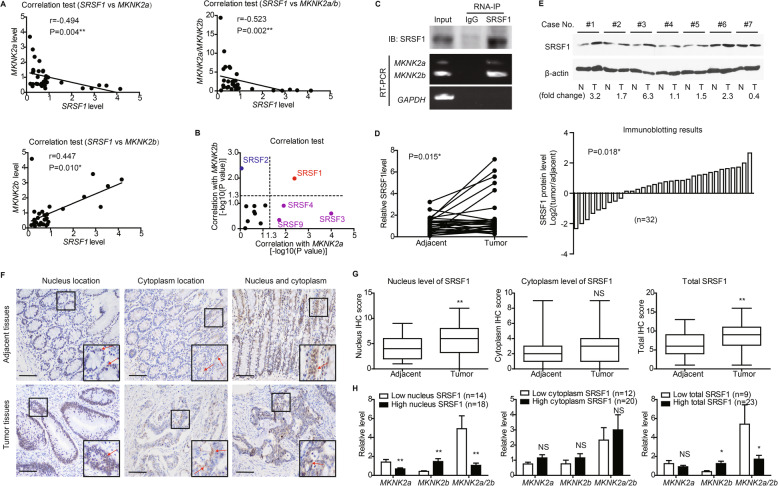
Fig. 2.
Correlation between nucleus SRSF1 location and MKNK2 alternative splicing. a SRSF1 level was tested by RT-qPCR in clinical CAC tissues (n = 32), and its correlations with MKNK2 variants were analyzed by Spearman correlation test. b RNA levels of other well-known splicing factors were also measured by RT-qPCR. Spearman correlation tests showed that only SRSF1 was significantly correlated with both MKNK2a and MKNK2b levels, indicating its potential effect on MKNK2a-MKNK2b switch. c SW480 cells transfected with pcDNA-vector or SRSF1 constructs were subjected to RNA-IP using anti-SRSF1 antibody in formaldehyde crosslinked nuclear extracts. The existence of SRSF1 protein and MKNK2-mRNA were tested via Western blotting and RT-PCR, respectively. d The mRNA levels of SRSF1 in 32 paired CAC tissues and adjacent nontumorous tissues were compared by RT-qPCR. P value was based on paired Student’s t-test. e Protein levels of SRSF1 in clinical specimens were also tested by western blotting (upper panel), and 71.9% cases (23/32) showed increased SRSF1 in tumor tissues (lower panel). f Immunohistochemical staining identified different subcellular location of SRSF1 in tissue specimens, including nucleus location and cytoplasm location. Scale bar: 100 μm. g Differences of SRSF1 IHC scores between CAC and adjacent nontumorous tissues were represented by box plots (n = 32). Top and bottom box edges represented the third and first quartile. Whiskers indicated highest and lowest score. P value was based on paired Student’s t-test. h By classifying patients into different subgroups according to ROC curves (Fig. S3D), we found that tissues with higher nucleus SRSF1 level or total SRSF1 level expressed more MKNK2b variant and showed a lower MKNK2a/2b ratio. In contrast, no statistical difference was observed based on cytoplasm SRSF1 level. P value was calculated by unpaired Student’s t-test