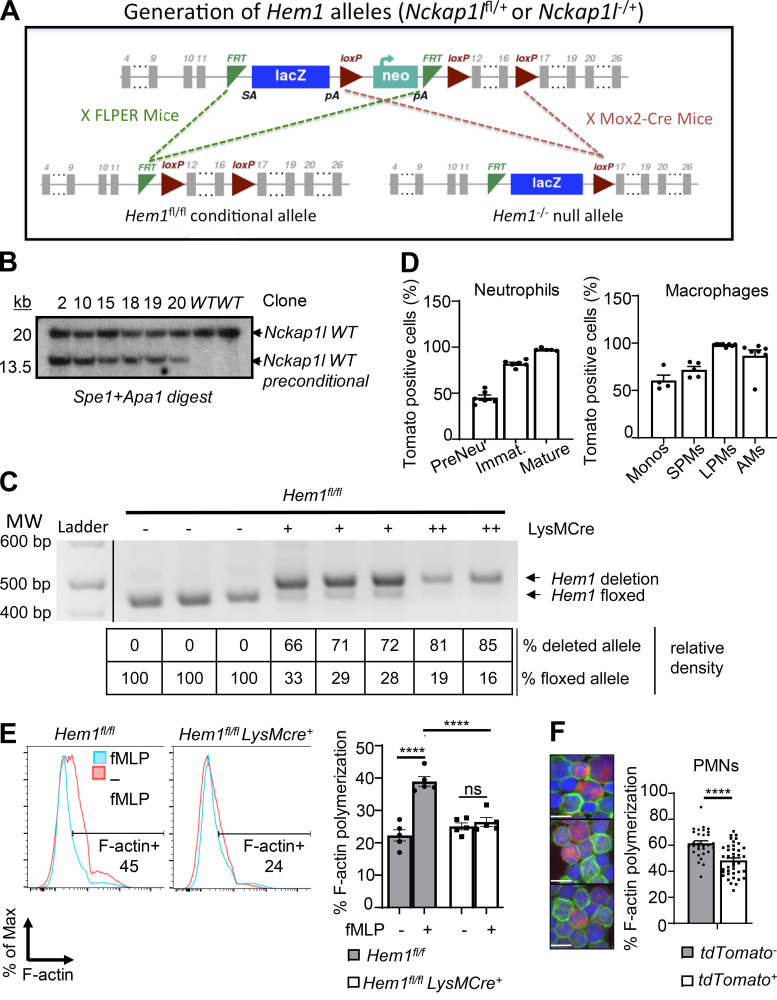
Figure S1.
Generation of constitutive and myeloid cell–specific Hem1-null mice. (A) Strategy used to generate Hem1 conditional mice. The Hem1fl/fl conditional targeting construct was generated by flanking exons 12–16 of the Nckap1l gene with loxP sites, with the addition of a lacZ reporter and neomycin selection cassette flanked by flippase recognition target (FRT) sites. Hem1 preconditional mice were bred to FLPER mice to delete the lacZ and Neo cassettes, resulting in Hem1 floxed conditional mice. Hem1−/− mice were generated by breeding Hem1 preconditional mice to Mox2-Cre mice, resulting in deletion of the Neo and FRT cassettes. (B) 6 ES cell clones were tested by Southern blotting and were confirmed to be correctly targeted to the Nckap1l locus. (C) PCR analysis of genomic DNA isolated from Hem1fl/flLysMCre+ BMDMs. Shown are the relative percentage of deleted versus floxed alleles based on semiquantitative densitometry. (D) Bar graphs show the percentages of RFP-positive cells reflective of Cre expression in the indicated populations of myeloid cells derived from Hem1fl/flLysMCre+tdTomato mice. (E) Flow cytometric histogram of BM neutrophils showing phalloidin staining (reflective of F-actin) with and without 1 μM fMLP stimulation for 2 min. Bar graph shows the percentage of phalloidin+ cells. (F) Fluorescence imaging of neutrophils from Hem1fl/flLysMCre+ tdTomato+ mice showing Cre-expressing cells (red), FITC phalloidin (green), and nuclei staining DAPI (blue; left) with bar graph showing the percentage of F-actin polymerization using ImageJ (right). Tomato+ cells have less F-actin polymerization (green). Scale bars, 10 µm. Data shown are mean ± SEM from n = 5–7 mice (6–12 wk old)/genotype representative of >2 independent exps. ****, P < 0.0001; Student’s two-tailed t test. Immat., immature; Monos, monocytes; PMNs, polymorphonuclear cells.