Abstract
Aims
Cell surface binding immunoglobin protein (csBiP) is predicted to be susceptible to SARS-CoV-2 binding. With a substrate-binding domain (SBD) that binds to polypeptides and a nucleotide-binding domain (NBD) that can initiate extrinsic caspase-dependent apoptosis, csBiP may be a promising therapeutic target for COVID-19. This study aims to identify FDA-approved drugs that can neutralize viral binding and prevent viral replication by targeting the functional domains of csBiP.
Methods
In silico screening of 1999 FDA-approved drugs against the functional domains of BiP were performed using three molecular docking programs to avoid bias from individual docking programs. Top ligands were selected by averaging the ligand rankings from three programs. Interactions between top ligands and functional domains of BiP were analyzed.
Key findings
The top 10 SBD-binding candidates are velpatasvir, irinotecan, netupitant, lapatinib, doramectin, conivaptan, fenoverine, duvelisib, irbesartan, and pazopanib. The top 10 NBD-binding candidates are nilotinib, eltrombopag, grapiprant, topotecan, acetohexamide, vemurafenib, paritaprevir, pixantrone, azosemide, and piperaquine-phosphate. Among them, Velpatasvir and paritaprevir are antiviral agents that target the protease of hepatitis C virus. Netupitant is an anti-inflammatory drug that inhibits neurokinin-1 receptor, which contributes to acute inflammation. Grapiprant is an anti-inflammatory drug that inhibits the prostaglandin E2 receptor protein subtype 4, which is expressed on immune cells and triggers inflammation. These predicted SBD-binding drugs could disrupt SARS-CoV-2 binding to csBiP, and NBD-binding drugs may falter viral attachment and replication by locking the SBD in closed conformation and triggering apoptosis in infected cells.
Significance
csBiP appears to be a novel therapeutic target against COVID-19 by preventing viral attachment and replication. These identified drugs could be repurposed to treat COVID-19 patients.
Keywords: COVID-19 / SARS-CoV-2, BiP, Drug repurposing, Molecular docking
Abbreviations: ACE2, angiotensin-converting enzyme 2; ARS, acute respiratory syndrome; BiP, binding immunoglobulin protein; csBiP, cell surface BiP; COVID-19, coronavirus disease 2019; EP4, PGE2 receptor protein subtype 4; ER, endoplasmic reticulum; HCV, hepatitis C virus; HSP, heat shock protein; MOA, mechanism of action; NBD, nucleotide binding domain; NK-1R, neurokinin-1 receptor; PDB, Protein Data Bank; PGE2, Prostaglandin E2; RdRp, RNA-dependent RNA polymerase; S protein, spike protein; SBD, substrate binding domain
Graphical abstract
1. Introduction
A novel strand of coronavirus, SARS-CoV-2, was reported in late 2019 (Bogoch et al., 2020; Li et al., 2020; Zhou et al., 2020). The disease caused by which was termed the coronavirus disease 2019 (COVID-19) (Naming the coronavirus disease, 2020). SARS-CoV-2 can utilize the angiotensin-converting enzyme 2 (ACE2) receptor to infiltrate the host cells (Lan et al., 2020; Wan et al., 2020; Liu et al., 2020; Chai et al., 2020)—the same receptor used by SARS-CoV (Li et al., 2003). The genome of SARS-CoV-2 closely resembles its predecessors: SARS-CoV in 2002 and MERS-CoV in 2012 (Chan et al., 2020; Ashour et al., 2020). SARS-CoV-2 has a higher binding affinity to ACE2, contributing to its amplified contagiousness (Lan et al., 2020; Shang et al., 2020; Letko et al., 2020). The mechanism of coronavirus entry has been reviewed by colleagues over the years (Hofmann and Pöhlmann, 2004; Belouzard et al., 2012; Cong and Ren, 2014; Fung and Liu, 2019; Hasan et al., 2020). Notably, coronaviruses can recognize many cell surface molecules for attachment. Among them, binding immunoglobulin protein (BiP), also known as Grp78 or HSPA5, emerged to be a common receptor with unique characteristics.
BiP was traditionally considered an endoplasmic reticulum (ER) chaperone that maintains ER homeostasis (Zhu and Lee, 2015). When cells are stressed such as in the conditions of viral infection, BiP is upregulated and trafficked to the plasma membrane (Gething, 1999; Alder and Johnson, 2004; Misra et al., 2005; Casas, 2017; Tsai and Lee, 2018). This phenomenon was observed clinically where COVID-19 patients had significantly higher expression of cell surface BiP (csBiP) in their alveolar epithelium (Palmeira et al., 2020; Aguiar et al., 2020). On the cell surface, BiP can facilitate viral attachment and entry such as for dengue virus (Jindadamrongwech et al., 2004; Chen et al., 2017), Japanese encephalitis virus (Nain et al., 2017), Zika virus (Pujhari et al., 2019; Royle et al., 2020), coxsackievirus (Triantafilou et al., 2002), Borna disease virus (Honda et al., 2009), Ebola virus (Reid et al., 2014), MERS-CoV, and bat coronavirus (Chu et al., 2018). A recent computational study predicted that csBiP can bind SARS-CoV-2 (Ibrahim et al., 2020). Therefore, as a crucial receptor protein in viral infection, csBiP deserves a closer look as a potential therapeutic target for COVID-19.
BiP has a substrate-binding domain (SBD) on the C-terminal region and a nucleotide-binding domain (NBD), also known as the ATPase domain, on the N-terminal region (Lindquist and Craig, 1988) (Fig. 1 A). The SBD binds to polypeptides often through hydrophobic interactions, which enables the chaperone duty of BiP (Rosenzweig et al., 2019). The NBD can alternate the SBD between open and closed conformations by ATP binding and hydrolysis (Yang et al., 2015). When BiP NBD is bound to ATP, the SBD has fast and dynamic binding and release of substrates; during ADP-bound state, the SBD has a closed conformation, preventing the exchange of substrates (Yang et al., 2015) (Fig. 1 B). When BiP NBD is inhibited and cannot bind to ATP, the SBD is locked into closed conformation (Yang et al., 2017). While neutralizing SARS-CoV-2 attachment by inhibiting BiP SBD has been proposed (Elfiky, 2020), targeting the NBD of BiP is hardly explored. Past investigations showed that antibody- and drug-inhibition of BiP NBD significantly reduced viral attachments (Jindadamrongwech et al., 2004; Chen et al., 2017; Nain et al., 2017; Pujhari et al., 2019; Royle et al., 2020; Triantafilou et al., 2002; Honda et al., 2009; Reid et al., 2014; Chu et al., 2018), warranting the significance of BiP NBD in viral studies. Furthermore, when the ATPase activity at the NBD of csBiP is disrupted, csBiP can trigger extrinsic caspase-dependent apoptosis pathways (Lanneau et al., 2008; Misra et al., 2009; Misra and Pizzo, 2010; Ge and Kao, 2019; Ko et al., 2015; Nair et al., 2014), providing a machinery to terminate infected cells. In this study, we aim to identify available drugs that could be repurposed to target csBiP SBD and NBD to interfere with SARS-CoV-2 binding and potentially induce apoptosis in the infected cells to prevent viral replication and further infection.
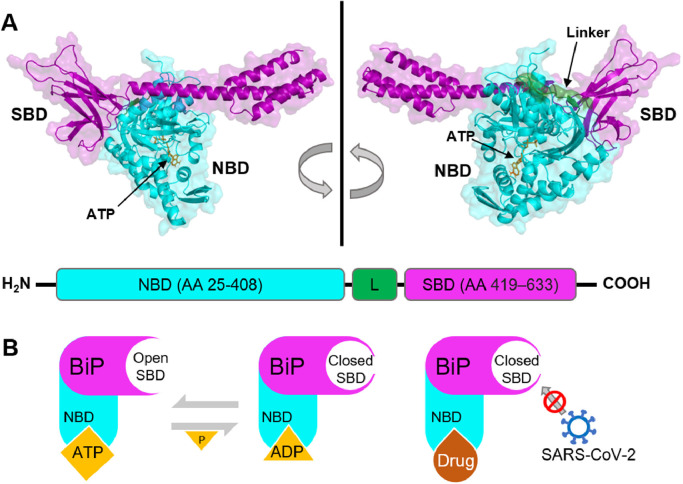
Fig. 1.
A. Full-length, ATP-bound BiP (PDB ID: 5E84) has two functional domains: a substrate-binding domain (SBD in violet) that binds to polypeptides and a nucleotide-binding domain (NBD in cyan) with ATPase activity. ATP is shown in orange sticks. A linker motif (L in green) connects the two domains. The NBD is at the N-terminus with amino acid (AA) residues 25 to 408. The SBD is at the C-terminus with AA residues 419 to 633. The linker connects the two domains and has AA residues 409 to 418. B. The SBD of BiP alternates between open and closed conformations, controlled by ATP binding and its hydrolysis. NBD inhibitors disrupt ATPase activity, locking the SBD at the closed conformation and preventing potential viral binding.
2. Materials and methods
2.1. Retrieval and preparation of ligands and protein receptor
The ligands were selected from the CLUE Repurposing Hub database (https://clue.io/repurposing-app) (Clue Repurposing, 2020), where all of the FDA approved drugs and drugs amid clinical trials are listed. Among them, the 1999 FDA-approved drugs were selected for virtual screening using three molecular docking programs: AutoDock4 (AD4) (Forli et al., 2016; Morris et al., 2009), AutoDock Vina (Vina) (Trott and Olson, 2010), and RosettaLigand (Rosetta) (DeLuca et al., 2015). The Simplified Molecular Input Line Entry System (SMILES) strings were obtained from the CLUE Repurposing Hub database for all 1999 drugs, and Schrodinger Maestro v 12.3 LigPrep module (Schrödinger Release, 2020) was used to prepare the drugs for molecular docking. LigPrep converted the SMILES strings to 3D SDF structures, desalted the drugs, and added all hydrogen atoms. Energy minimization was performed on the drugs using the OPLS3e force field (Harder et al., 2016). The 1999 ligand library in SDF format was used to generate a conformer library, which was needed for docking with Rosetta, using the Conformer Generation tool from the BCL suite (Kothiwale et al., 2015). OpenBabel software (O'Boyle et al., 2011) was used to convert the ligands in SDF format to PDBQT format with Gasteiger charges added, which was needed for docking with AD4 and Vina. The full-length and wild-type 3D crystal structure of BiP (PDB ID: 5E84, Resolution: 2.99 Å) was retrieved from the Protein Data Bank (PDB) (Yang et al., 2015). For molecular docking in AD4 and Vina, the crystal structure of BiP monomer was loaded into AutoDock Tools software (Morris et al., 2009) for preparation: polar hydrogen atoms were added; water molecules, ion molecules, and bound ATP were removed; and lastly, Kollman Charges were added. Prepared BiP was output in PDBQT format for molecular docking in Vina and AD4. For molecular docking in Rosetta, the BiP PDB file was prepared using Rosetta's clean PDB script (Lemmon and Meiler, 2012).
2.2. Molecular docking and analysis
For molecular docking using AutoDock 4 software (Forli et al., 2016; Morris et al., 2009) and AutoDock Vina software (Trott and Olson, 2010), the docking was performed in the high-performance computing facility at the University of Alabama at Birmingham. For molecular docking using RosettaLigand software (DeLuca et al., 2015), the docking was performed in high performance supercomputer in Alabama Supercomputer Authority. The selected 1999 FDA-approved drugs were the ligands, and the NBD and the SBD of BiP (PDB ID: 5E84) were docking targets. ATP and pep42 were chosen as reference substrates and positive controls for the NBD and SBD respectively because they are known NBD- and SBD-binding ligands (Kim et al., 2006; Liu et al., 2007). In molecular docking, the ligands were treated as flexible with rotatable bonds while the receptor protein BiP was treated as rigid. Docking results were first ranked based on each individual molecular docking program according to the calculated binding energy (kcal/mol) in Vina and AD4 and the interface energy (Rosetta Energy Units (REU)) in Rosetta. After the drugs were ranked based on the individual programs, the average of the ranks from three docking programs was calculated for each ligand to avoid the bias of each individual docking program, and the averages were used to rank the drugs’ overall promise.
Drug compounds with high binding affinities (low negative binding energy) to the NBD or the SBD were selected as the NBD- or SBD-inhibitor candidates. Hydrophobic interactions, hydrogen bond formations, and electrostatic interactions between BiP (PDB ID: 5E84) and the identified drugs were performed and visualized using PyMol (Schrodinger, LLC., 2015) and Schrödinger Maestro (Maestro, 2020) software.
2.3. Electrostatic potential analysis
To evaluate the surface electrostatic potential of BiP in complex with the identified top drug candidates, we generated electrostatic potential maps using adaptive Poisson-Boltzmann solver (APBS) plugin function in PyMOL software (Lerner and Carlson, 2020). The APBS plugin in PyMOL is an interface of APBS program (Baker et al., 2001) in PyMol and provides easy access to electrostatics calculations and the visualization of the electrostatic potential on protein surfaces.
3. Results and discussion
3.1. Available drugs repurposed to target the SBD of BiP
Although without available crystallographic evidence of SARS-CoV-2-bound BiP, a recent in silico modeling study suggested that the Cys480-Cys488 region of SARS-CoV-2 S protein could bind to the SBD of BiP (active residues Thr428, Phe451, and Ser452) (Ibrahim et al., 2020). Subsequent molecular docking studies proposed using natural ligands such as phytoestrogens and estrogens (Elfiky, 2020) and peptidic drugs such as Zilucoplan, Obinepitide, and Corticorelin ovine triflutate (Palmeira et al., 2020) to compete with viral S protein for BiP-binding. In this study, we screened FDA-approved small molecular drugs that could be repurposed to target BiP.
Pep42 is a known peptidic substrate of BiP SBD (Kim et al., 2006; Liu et al., 2007) and is used as a reference and positive control for tested drug candidates. The top 10 SBD-binding ligands are velpatasvir, irinotecan, netupitant, lapatinib, doramectin, conivaptan, fenoverine, duvelisib, irbesartan, and pazopanib (Table 1 ). Velpatasvir (-8.1 kcal/mol in Vina; -7.9 kcal/mol in AD4; -12.2 REU in Rosetta) has the strongest overall binding to BiP SBD (Table 1 , Fig. 2 A), forming hydrogen bonds with residues Thr434, Lys435, and Gln449 (Fig. 2 B). With partial negative charges in functional groups such as hydroxyl, ester, ether, and amide in velpatasvir, the two ends of velpatasvir electrostatically interact with positively charged residues on the exterior of BiP SBD such as Lys 435, Lys 447, and Lys 460, (Fig. 2 C and D). Its cyclohexane backbone exhibits hydrophobic interaction with Ile 426, Val 429, Phe 451, and Val 453 (Fig. 2 D).
Table 1.
The molecular docking results of top 10 drug candidates that may be repurposed to prevent SARS-CoV-2 from binding to the SBD of BiP.
| Compound Name | Structure | Category | Mechanism of Action (MOA) | Vina Binding Energy (kcal/mol) | AD4 Binding Energy (kcal/mol) | Rosetta Interface Energy (REU) | Overall Ranking for FDA-approved drug |
|---|---|---|---|---|---|---|---|
| Pep42 |  |
Positive Control | BiP SBD ligand (Kim et al., 2006) | -5.8 | -8.5 | -5.0 | |
| Velpatasvir | 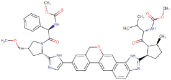 |
Antiviral | HCV NS5A inhibitor | -8.1 | -7.9 | -12.2 | 1 |
| Irinotecan |  |
Anticancer | Topoisomerase I inhibitor | -7.8 | -7.8 | -12.0 | 2 |
| Netupitant | 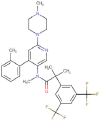 |
Antiemitic | NK-1 receptor antagonist | -8.6 | -7.9 | -11.4 | 3 |
| Lapatinib |  |
Anticancer | EGFR/HER2 protein kinase inhibitor | -7.0 | -7.6 | -12.1 | 4 |
| Doramectin |  |
Antiparasitic | Unknown | -7.5 | -9.3 | -10.7 | 5 |
| Conivaptan | 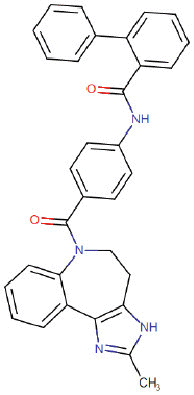 |
Cardiovascular | Antagonist of human arginine vasopressin | -8.6 | -7.4 | -11.1 | 6 |
| Fenoverine | 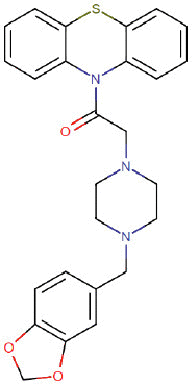 |
Gastro-intestinal | Acetylcholine receptor antagonist | -7.2 | -7.2 | -11.9 | 7 |
| Duvelisib | 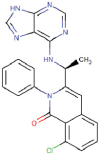 |
Anticancer | Phosphoinositide-3 kinases inhibitor | -7.0 | -8.0 | -11.4 | 8 |
| Irbesartan | 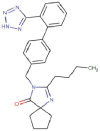 |
Cardiovascular | Type-1 angiotensin II receptor antagonist | -7.3 | -6.9 | -12.0 | 9 |
| Pazopanib |  |
Anticancer | Tyrosine kinase inhibitor | -6.9 | -6.9 | -13.1 | 10 |
Fig. 2.
Interaction between velpatasvir (shown in orange sticks) and the SBD of BiP. A. Predicted docking of velpatasvir inside the substrate binding site of the BiP SBD (shown in violet). The NBD is indicated in cyan. B. H-bonds between velpatasvir and residues Thr 434, Lys 435, and Gln 449 are illustrated by light green dashed lines in three-dimensional space. C. Electrostatic interaction between velpatasvir and the SBD of BiP. Red: negative electrostatic potential; Blue: positive electrostatic potential; White: neutral electrostatic potential. D. 2D schematic representation of interactions between velpatasvir and active residues. Atoms with an electric charge greater than +0.15 or less than -0.15 coulombs (C) are labeled. H-bonds are denoted by purple arrows. Hydrophobic interactions are denoted by green ribbons between BiP residues and velpatasvir. The grey rings around the ligand atoms indicate the solvent exposure.
Velpatasvir is an antiviral agent often administered with sofosbuvir under the brand name Epclusa to treat hepatitis C virus (HCV) infection—another single-stranded RNA virus (Greig, 2016). Numerous computational studies have implicated velpatasvir as a potential inhibitor of SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) (Indu et al., 2020; Bello et al., 2020; Chen et al., 2020). Results from our study suggest that velpatasvir may compete with SARS-CoV-2 for binding the SBD of BiP, thereby preventing viral attachment.
Netupitant, on the other hand, is a less obvious but highly promising drug candidate in combating COVID-19. It also demonstrated strong binding to BiP SBD (-8.6 kcal/mol in Vina; -7.9 kcal/mol in AD4; -11.4 REU in Rosetta) (Table 1, Fig. 3 A). The partial negative charges due to amide groups on netupitant electrostatically interact with positively charged exterior regions of BiP SBD (Fig. 3 C and D). Meanwhile, its non-polar ring structure sits deep inside the SBD through hydrophobic interaction with nonpolar residues such as Ile 426, Val 429, Ile 456, Phe 451, Val 453, and Val 457 (Fig. 3 B and D).
Fig. 3.
Interaction between netupitant (shown in orange sticks) and the SBD of BiP. A. Predicted docking of netupitant inside the substrate binding site of the BiP SBD (shown in violet). B. Close up look of netupitant inside the SBD of BiP through hydrophobic interactions. C. Electrostatic interaction between netupitant and the SBD of BiP. Red: negative electrostatic potential; Blue: positive electrostatic potential; White: neutral electrostatic potential. D. 2D schematic representation of interactions between netupitant and active residues. Atoms with an electric charge greater than +0.15 or less than -0.15 coulombs (C) are labeled. Hydrophobic interactions are denoted by green ribbons between BiP residues and velpatasvir. The grey rings around the ligand atoms indicate the solvent exposure.
Netupitant is an antagonist of the neurokinin-1 receptor (NK-1R), a G protein-coupled receptor that is normally activated by Substance P (SP). Upon SP binding, NK-1R initiates signaling cascades to induce transcription factor NF-κb and subsequent proinflammatory cytokines (Williams et al., 2007; Bost, 2004). During viral infection and inflammation, NK-1R is upregulated on the surface of pulmonary epithelial cells and immune cells (Bai et al., 1995; Chu et al., 2000; King et al., 2001). Meanwhile, SP is produced and released into the extracellular space by airway epithelia to escalate the inflammation through autocrine and paracrine signaling (King et al., 2001; Stewart et al., 2008; Suvas, 2017). This positive-feedback inflammatory machinery likely contributes to the clinically observed acute respiratory syndromes (ARSs) in COVID-19 patients. To treat COVID-19 symptoms, a clinical study in Pakistan used another NK-1R antagonist, aprepitant (Mehboob et al., 2020). The positive results from this study implicated the promise of NK-1R antagonists as a therapeutic candidate for COVID-19. Our data suggest that netupitant may also inhibit the SBD of BiP. Therefore, in addition to the anti-inflammatory effects of netupitant, we herein suggest a novel strategy whereby netupitant may prevent SARS-CoV-2 from attaching to csBiP.
3.2. Available drugs repurposed to target the NBD of BiP
ATP, the natural substrate of BiP NBD, is used as a reference and positive control for drug candidates. The top 10 ligands are nilotinib, eltrombopag, grapiprant, topotecan, acetohexamide, vemurafenib, paritaprevir, pixantrone, azosemide, and piperaquine-phosphate (Table 2 ). While nilotinib (-9.7 kcal/mol in Vina; -6.6 kcal/mol in AD4; -20.1 REU in Rosetta) and eltrombopag (-10.7 kcal/mol in Vina; -7.1 kcal/mol in AD4; -13.8 REU in Rosetta) demonstrated slightly better overall binding against BiP NBD (Table 2), they both have risk factors associated with cytotoxicity and hepatoxicity (Hochhaus et al., 2016; Sadiq et al., 2019). Grapiprant presents the least toxicity (Kirkby Shaw et al., 2015) and greatest potential to be a drug candidate for COVID-19. It demonstrated strong binding affinity (-9.3 kcal/mol in Vina; -6.5 kcal/mol in AD4; -16.2 REU in Rosetta) to the NBD of BiP (Table 2 , Fig. 4 A) by forming hydrogen bonds with residues Thr 38 and Glu 256 (Fig. 4 B and D). The partial negative charges on grapiprant due to amide and hydroxyl groups fit in close proximity with positively charged regions of BiP NBD by electrostatically interacting with positively charged residues such as Arg 289, Arg 290, Lys 294, Lys 296, and Arg 297 (Fig. 4 C and D).
Table 2.
The molecular docking results of top 10 drug candidates that may be repurposed to inhibit the NBD of BiP thereby disrupting its ATPase functions.
| Compound Name | Structure | Category | Mechanism of Action (MOA) | Vina Binding Energy (kcal/mol) | AD4 Binding Energy (kcal/mol) | Rosetta Interface Energy (REU) | Overall Ranking for FDA-approved drug |
|---|---|---|---|---|---|---|---|
| ATP |  |
Control | Natural BiP NBD Ligand | -8.8 | -2.7 | -9.1 | |
| Nilotinib |  |
Anticancer | Tyrosine kinase inhibitor | -9.7 | -6.6 | -20.1 | 1 |
| Eltrombopag | 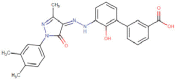 |
Cardiovascular | Thrombopoietin receptor agonist | -10.7 | -7.1 | -13.8 | 2 |
| Grapiprant |  |
Anti-inflammatory | Prostaglandin EP4 receptor inhibitor | -9.3 | -6.5 | -16.2 | 3 |
| Topotecan | 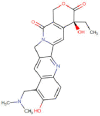 |
Anticancer | Topoisomerase I inhibitor | -8.8 | -6.8 | -15.7 | 4 |
| Acetohexamide | 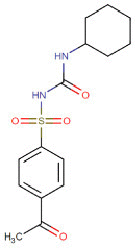 |
Diabetic | ATP-dependent K+ channel inhibitor | -9.2 | -6.9 | -13.8 | 5 |
| Vemurafenib | 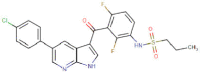 |
Anticancer | Protein kinase inhibitor | -10.1 | -7.2 | -13.0 | 6 |
| Paritaprevir | 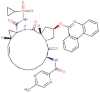 |
Antiviral | NS3/4A serine protease inhibitor | -9.2 | -7.2 | -13.2 | 7 |
| Pixantrone |  |
Anticancer | DNA-topoisomerase II complex stabilizer | -8.8 | -6.7 | -14.4 | 8 |
| Azosemide |  |
Cardiovascular | Electrolyte reabsorption inhibitor | -8.7 | -6.6 | -15.3 | 9 |
| Piperaquine-phosphate |  |
Antiparasitic | Unknown | -9.4 | -6.9 | -12.8 | 10 |
Fig. 4.
Interaction between grapiprant (shown in orange sticks) and the NBD of BiP. A. Predicted docking of grapiprant inside the ATP-binding pocket of BiP NBD (shown in cyan). The NBD is indicated in cyan. B. H-bonds between grapiprant and residues Thr38 and Glu256 are illustrated by light green dashed lines in three-dimensional space. C. Electrostatic interaction between grapiprant and the NBD of BiP. Red: negative electrostatic potential; Blue: positive electrostatic potential; White: neutral electrostatic potential. D. 2D schematic representation of interactions between grapiprant and active residues. Atoms with an electric charge greater than +0.15 or less than -0.15 coulombs (C) are labeled. H-bonds are denoted by purple arrows. The grey rings around the ligand atoms indicate the solvent exposure.
Grapiprant is an anti-inflammatory prostaglandin inhibitor currently used to treat inflammatory pain by competitively inhibiting the prostaglandin E2 (PGE2) receptor protein subtype 4 (EP4) (Nakao et al., 2007). EP4 is abundantly expressed on the surface of lung cells and almost all immune cells (Robb et al., 2020). After PGE2-binging, EP4 can initiate a wide range of physiological effects including cellular differentiation, proliferation, angiogenesis, and inflammation (Kalinski, 2012; Konya et al., 2013). In patients infected with SARS-CoV-1 and SARS-CoV -2, PGE2 level has been shown to be upregulated (Lee et al., 2004; Hong et al., 2020). A past study on influenza A virus also showed that PGE2 can undermine the host antiviral response by inhibiting both innate and adaptive immunity (Coulombe et al., 2014), and a recent study suggests PGE2 may be responsible for early inflammation in COVID-19 patients (Robb et al., 2020). Therefore, as an inhibitor to the receptor of PGE2, grapiprant may help fight infection and ameliorate COVID-19 symptoms. Corroborating with this idea, the results from our study implicate that grapiprant could inhibit the ATPase activity of csBiP and allosterically lock the SBD in the closed conformation (illustrated in Fig. 1 B). The locked SBD conformation may prevent SARS-CoV-2 from binding to csBiP, while disrupted ATPase activity may trigger extrinsic caspase-dependent apoptosis (Lanneau et al., 2008; Misra et al., 2009; Misra and Pizzo, 2010; Ge and Kao, 2019; Ko et al., 2015; Nair et al., 2014), a process analogous to that of cytotoxic T cell-initiated cellular immunity (Actor, 2012). This way, upregulated csBiP due to SARS-CoV-2 infection could be employed as a therapeutic target to fight against the virus and remedy the disease.
In addition to grapiprant, paritaprevir showed strong binding with the NBD of BiP (-9.2 kcal/mol in Vina; -7.2 kcal/mol in AD4; -13.2 REU in Rosetta) as well (Table 2 , Fig. 5 A). Hydrogen bonding exists between its hydroxyl group and BiP residue Arg 297, which “hooks” paritaprevir to the binding pocket of BiP NBD (Fig. 5 B and D). Meanwhile, the partially negative amide group on the ring electrostatically interacts with positively charged residues such as Lys 294, Lys 296, Arg 297, and Arg 367, further locking the three connected ring structure inside the ATP-binding pocket (Figure 5 C and 5 D). Similar to velpatasvir, paritaprevir is an antiviral agent against the HCV protease complex that gained a considerable amount of attention in the studies of COVID-19. Through tools like artificial intelligence (AI) deep learning, molecular docking, and, in some studies, molecular dynamics simulation, these investigations unanimously suggested the potential of paritaprevir in inhibiting viral RdRp, S protein, and proteases (Manikyam and Joshi, 2020; Shah et al., 2020; Alamri et al., 2020; Khan et al., 2020; Choi et al., 2020). Our results expand the current paradigm and suggest that paritaprevir may also be used to target the NBD of csBiP, thereby sabotaging viral activities.
Fig. 5.
Interaction between paritaprevir (shown in orange sticks) and the NBD of BiP. A. Predicted docking of paritaprevir inside the ATP-binding pocket of BiP NBD (shown in cyan). The NBD is indicated in cyan. B. H-bond between paritaprevir and residues Arg297 is illustrated by light green dashed lines in three-dimensional space. C. Electrostatic interaction between paritaprevir and the NBD of BiP. Red: negative electrostatic potential; Blue: positive electrostatic potential; White: neutral electrostatic potential. D. 2D schematic representation of interactions between paritaprevir and active residues. Atoms with an electric charge greater than +0.15 or less than -0.15 coulombs (C) are labeled. H-bond is denoted by purple arrows. The grey rings around the ligand atoms indicate the solvent exposure.
Notably, it is by no coincidence that there are numerous anti-cancer drugs among top-ranked drugs for both BiP SBD and NBD (Tables 1 & 2). Since csBiP can initiate caspase-dependent apoptosis, it has become a popular target for cancer therapies (Casas, 2017; Ge and Kao, 2019; Gopal and Pizzo, 2018; Araujo et al., 2018). However, anti-cancer drugs may bring complications beyond COVID-19 symptoms and, therefore, are not discussed as therapeutic candidates against SARS-CoV-2 infection.
In summary, we hereby propose a novel therapeutic strategy against COVID-19 by targeting the two functional domains of csBiP. Since the SBD of csBiP is susceptible to viral binding, the drugs that are predicted to inhibit BiP SBD may prevent SARS-CoV-2 attachment (Table 1 and Supplemental Table 1). Meanwhile, the NBD of csBiP not only governs the allosteric conformation of SBD but also possesses pro-apoptotic functions. Therefore, the drugs that are predicted to inhibit BiP NBD may trigger apoptosis in infected cells, prevent viral attachment and replication (Table 2 and Supplemental Table 2). Among the drugs, velpatasvir, netupitant, grapiprant, and paritaprevir are the most promising candidates because of their original modes of action and calculated strong binding with BiP.
3. Conclusion
Despite the efforts worldwide in elucidating the pathophysiology of COVID-19, an effective treatment is still desperately lacking. csBiP can act as a receptor that enables pathogen attachment and also initiate caspase-dependent apoptosis pathways when its ATPase activity is disrupted. Using in silico screening approach, we identified drugs that demonstrated high binding affinities with two functional domains of BiP. Among the drug candidates, velpatasvir and netupitant may interfere with SARS-CoV-2 attachment by inhibiting the SBD of BiP; grapiprant and paritaprevir may lock the SBD conformation to a closed state and trigger extrinsic caspase-dependent apoptosis by disrupting the ATPase activity of BiP NBD. The known anti-inflammatory modes of action by netupitant and grapiprant may palliate the acute pulmonary immune response and mitigate COVID-19 symptoms; the known antiviral function of velpatasvir and paritaprevir may inhibit rival RdRp and prevent SARS-CoV-2 replication. Our results expand the current paradigm and suggest that the identified drugs could be used to target csBiP, thereby sabotaging viral activities. Results from this study could inspire the future in vitro, in vivo, and clinical trials towards available drugs being repurposed to treat COVID-19 patients.
Data availability
The docking structures are available upon request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Author contributions
Yiming Zhang: Conceptualization, Investigation, Data curation, Formal analysis, Writing, Visualization; Rory A Greer: Data curation, Formal analysis, Writing; Yuwei Song: Data curation, Visualization, Formal analysis, Writing; Hrithik Praveen: Data curation, Validation, Writing; Yuhua Song: Conceptualization, Methodology, Writing, Supervision, Project administration.
Declaration of Competing Interest
The authors declare no competing interest in this work.
Acknowledgments
This work is funded by the University of Alabama at Birmingham Honors College Presidential Summer Fellowship (YZ) and pilot grant from Hugh Kaul Precision Medicine Institute at University of Alabama at Birmingham (YHS). This work is enabled by the high-performance computing recourses at UAB IT Research Computing and at Alabama Supercomputer Authority.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ejps.2021.105771.
Appendix. Supplementary materials
References
- Actor J.K. In: Elsevier's Integrated Review Immunology and Microbiology (Second Edition) Actor J.K., editor. W.B. Saunders; Philadelphia: 2012. 4 - T-cell immunity; pp. 25–32. [Google Scholar]
- Aguiar, J.A., Tremblay, B.J.M., Mansfield, M.J., Woody, O., Lobb, B., Banerjee, A., Chandiramohan, A., Tiessen, N., Dvorkin-Gheva, A., Revill, S., Miller, M.S., Carlsten, C., Organ, L., Joseph, C., John, A., Hanson, P., McManus, B.M., Jenkins, G., Mossman, K., Ask, K., Doxey, A.C., and Hirota, J.A. (2020) Gene expression and <em>in situ</em> protein profiling of candidate SARS-CoV-2 receptors in human airway epithelial cells and lung tissue, bioRxiv, 2020.2004.2007.030742. [DOI] [PMC free article] [PubMed]
- Alamri M.A., Tahir Ul Qamar M., Mirza M.U., Bhadane R., Alqahtani S.M., Muneer I., Froeyen M., Salo-Ahen O.M.H. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CL(pro) J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1782768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder N.N., Johnson A.E. Cotranslational membrane protein biogenesis at the endoplasmic reticulum. J. Biol. Chem. 2004;279:22787–22790. doi: 10.1074/jbc.R400002200. [DOI] [PubMed] [Google Scholar]
- Araujo N., Hebbar N., Rangnekar V.M. GRP78 Is a targetable receptor on cancer and stromal cells. EBioMedicine. 2018;33:2–3. doi: 10.1016/j.ebiom.2018.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashour H.M., Elkhatib W.F., Rahman M.M., Elshabrawy H.A. Insights into the recent 2019 novel coronavirus (SARS-CoV-2) in light of past human coronavirus outbreaks. Pathogens (Basel, Switzerland) 2020;9 doi: 10.3390/pathogens9030186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai T.R., Zhou D., Weir T., Walker B., Hegele R., Hayashi S., McKay K., Bondy G.P., Fong T. Substance P (NK1)- and neurokinin A (NK2)-receptor gene expression in inflammatory airway diseases. Am. J. Physiol. 1995;269:L309–L317. doi: 10.1152/ajplung.1995.269.3.L309. [DOI] [PubMed] [Google Scholar]
- Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello M., Martínez-Muñoz A., Balbuena-Rebolledo I. Identification of saquinavir as a potent inhibitor of dimeric SARS-CoV2 main protease through MM/GBSA. J. Mol. Model. 2020;26:340. doi: 10.1007/s00894-020-04600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Millet J.K., Licitra B.N., Whittaker G.R. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. doi: 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogoch I.I., Watts A., Thomas-Bachli A., Huber C., Kraemer M.U.G., Khan K. Pneumonia of unknown aetiology in Wuhan, China: potential for international spread via commercial air travel. J. Travel Med. 2020;27 doi: 10.1093/jtm/taaa008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost K.L. Tachykinin-mediated modulation of the immune response. Front. Biosci.: J. Virtual Lib. 2004;9:3331–3332. doi: 10.2741/1484. [DOI] [PubMed] [Google Scholar]
- Casas C. GRP78 at the centre of the stage in cancer and neuroprotection. Front. Neurosci. 2017;11:177. doi: 10.3389/fnins.2017.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai, X., Hu, L., Zhang, Y., Han, W., Lu, Z., Ke, A., Zhou, J., Shi, G., Fang, N., Fan, J., Cai, J., Fan, J., and Lan, F.. (2020) Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection, bioRxiv, 2020.2002.2003.931766.
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.H., Chen C.C., Lin Y.S., Chang P.C., Lu Z.Y., Lin C.F., Chen C.L., Chang C.P. AR-12 suppresses dengue virus replication by down-regulation of PI3K/AKT and GRP78. Antiviral Res. 2017;142:158–168. doi: 10.1016/j.antiviral.2017.02.015. [DOI] [PubMed] [Google Scholar]
- Chen Y.W., Yiu C.B., Wong K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research. 2020;9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Shin B., Kang K., Park S., Beck B.R. Target-centered drug repurposing predictions of human angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine subtype 2 (TMPRSS2) interacting approved drugs for coronavirus disease 2019 (COVID-19) treatment through a drug-target interaction deep learning model. Viruses. 2020;12 doi: 10.3390/v12111325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan C.M., Zhang X., Wang Y., Yuan S., Zhou J., Au-Yeung R.K., Sze K.H., Yang D., Shuai H., Hou Y., Li C., Zhao X., Poon V.K., Leung S.P., Yeung M.L., Yan J., Lu G., Jin D.Y., Gao G.F., Chan J.F., Yuen K.Y. Middle East respiratory syndrome coronavirus and bat coronavirus HKU9 both can utilize GRP78 for attachment onto host cells. J. Biol. Chem. 2018;293:11709–11726. doi: 10.1074/jbc.RA118.001897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.W., Kraft M., Krause J.E., Rex M.D., Martin R.J. Substance P and its receptor neurokinin 1 expression in asthmatic airways. J. Allergy Clin. Immunol. 2000;106:713–722. doi: 10.1067/mai.2000.109829. [DOI] [PubMed] [Google Scholar]
- (2020) Clue Repurposing, Broad institute, clue the drug repurposing hub.
- Cong Y., Ren X. Coronavirus entry and release in polarized epithelial cells: a review. Rev. Med. Virol. 2014;24:308–315. doi: 10.1002/rmv.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe F., Jaworska J., Verway M., Tzelepis F., Massoud A., Gillard J., Wong G., Kobinger G., Xing Z., Couture C., Joubert P., Fritz JH., Powell WS., Divangahi M. Targeted prostaglandin E2 inhibition enhances antiviral immunity through induction of type I Interferon and apoptosis in macrophages. Immunity. 2014;40:554–568. doi: 10.1016/j.immuni.2014.02.013. [DOI] [PubMed] [Google Scholar]
- DeLuca S., Khar K., Meiler J. Fully flexible docking of medium sized ligand libraries with RosettaLigand. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky A.A. Natural products may interfere with SARS-CoV-2 attachment to the host cell. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1761881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forli S., Huey R., Pique M.E., Sanner M.F., Goodsell D.S., Olson A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Human coronavirus: host-pathogen interaction. Annu. Rev. Microbiol. 2019;73:529–557. doi: 10.1146/annurev-micro-020518-115759. [DOI] [PubMed] [Google Scholar]
- Ge R., Kao C. Cell surface GRP78 as a death receptor and an anticancer drug target. Cancers (Basel) 2019;11 doi: 10.3390/cancers11111787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M.J. Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- Gopal, U., and Pizzo, S.V. (2018) The endoplasmic reticulum chaperone GRP78 also functions as a cell surface signaling receptor, 9-40.
- Greig S.L. Sofosbuvir/velpatasvir: a review in chronic hepatitis C. Drugs. 2016;76:1567–1578. doi: 10.1007/s40265-016-0648-2. [DOI] [PubMed] [Google Scholar]
- Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y., Wang L., Lupyan D., Dahlgren M.K., Knight J.L., Kaus J.W., Cerutti D.S., Krilov G., Jorgensen W.L., Abel R., Friesner R.A. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- Hasan A., Paray B.A., Hussain A., Qadir F.A., Attar F., Aziz F.M., Sharifi M., Derakhshankhah H., Rasti B., Mehrabi M., Shahpasand K., Saboury A.A., Falahati M. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochhaus A., Saglio G., Hughes T.P., Larson R.A., Kim D.W., Issaragrisil S., le Coutre P.D., Etienne G., Dorlhiac-Llacer P.E., Clark R.E., Flinn I.W., Nakamae H., Donohue B., Deng W., Dalal D., Menssen H.D., Kantarjian H.M. Long-term benefits and risks of frontline nilotinib vs imatinib for chronic myeloid leukemia in chronic phase: 5-year update of the randomized ENESTnd trial. Leukemia. 2016;30:1044–1054. doi: 10.1038/leu.2016.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pöhlmann S. Cellular entry of the SARS coronavirus. Trends Microbiol. 2004;12:466–472. doi: 10.1016/j.tim.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda T., Horie M., Daito T., Ikuta K., Tomonaga K. Molecular chaperone BiP interacts with Borna disease virus glycoprotein at the cell surface. J. Virol. 2009;83:12622–12625. doi: 10.1128/JVI.01201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W., Chen Y., You K., Tan S., Wu F., Tao J., Chen X., Zhang J., Xiong Y., Yuan F., Yang Z., Chen T., Chen X., Peng P., Tai Q., Wang J., Zhang F., Li Y.-X. Celebrex adjuvant therapy on coronavirus disease 2019: an experimental study. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.561674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J. Infect. 2020;80:554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indu P., Rameshkumar M.R., Arunagirinathan N., Al-Dhabi N.A., Valan Arasu M., Ignacimuthu S. Raltegravir, Indinavir, Tipranavir, Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: a molecular docking and drug repurposing approach. J. Infect. Public Health. 2020;13:1856–1861. doi: 10.1016/j.jiph.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindadamrongwech S., Thepparit C., Smith D.R. Identification of GRP 78 (BiP) as a liver cell expressed receptor element for dengue virus serotype 2. Arch. Virol. 2004;149:915–927. doi: 10.1007/s00705-003-0263-x. [DOI] [PubMed] [Google Scholar]
- Kalinski P. Regulation of immune responses by prostaglandin E2. J. Immunol. 2012;188:21–28. doi: 10.4049/jimmunol.1101029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan R.J., Jha R.K., Singh E., Jain M., Amera G.M., Singh R.P., Muthukumaran J., Singh A.K. Identification of promising antiviral drug candidates against non-structural protein 15 (NSP15) from SARS-CoV-2: an in silico assisted drug-repurposing study. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1814870. [DOI] [PubMed] [Google Scholar]
- Kim Y., Lillo A.M., Steiniger S.C.J., Liu Y., Ballatore C., Anichini A., Mortarini R., Kaufmann G.F., Zhou B., Felding-Habermann B., Janda K.D. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45:9434–9444. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- King K.A., Hu C., Rodriguez M.M., Romaguera R., Jiang X., Piedimonte G. Exaggerated neurogenic inflammation and substance P receptor upregulation in RSV-infected weanling rats. Am. J. Respir. Cell Mol. Biol. 2001;24:101–107. doi: 10.1165/ajrcmb.24.2.4264. [DOI] [PubMed] [Google Scholar]
- Kirkby Shaw K., Rausch-Derra L.C., Rhodes L. Grapiprant: an EP4 prostaglandin receptor antagonist and novel therapy for pain and inflammation. Vet. Med. Sci. 2015;2:3–9. doi: 10.1002/vms3.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S.K., Kim J., Na D.C., Park S., Park S.H., Hyun J.Y., Baek K.H., Kim N.D., Kim N.K., Park Y.N., Song K., Shin I. A small molecule inhibitor of ATPase activity of HSP70 induces apoptosis and has antitumor activities. Chem. Biol. 2015;22:391–403. doi: 10.1016/j.chembiol.2015.02.004. [DOI] [PubMed] [Google Scholar]
- Konya V., Marsche G., Schuligoi R., Heinemann A. E-type prostanoid receptor 4 (EP4) in disease and therapy. Pharmacol. Ther. 2013;138:485–502. doi: 10.1016/j.pharmthera.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothiwale S., Mendenhall J.L., Meiler J. BCL::Conf: small molecule conformational sampling using a knowledge based rotamer library. J. Cheminform. 2015;7:47. doi: 10.1186/s13321-015-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lanneau D., Brunet M., Frisan E., Solary E., Fontenay M., Garrido C. Heat shock proteins: essential proteins for apoptosis regulation. J. Cell Mol. Med. 2008;12:743–761. doi: 10.1111/j.1582-4934.2008.00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H., Chen R.-F., Liu J.-W., Yeh W.-T., Chang J.-C., Liu P.-M., Eng H.-L., Lin M.-C., Yang K.D. Altered p38 mitogen-activated protein kinase expression in different leukocytes with increment of immunosuppressive mediators in patients with severe acute respiratory syndrome. J. Immunol. 2004;172:7841. doi: 10.4049/jimmunol.172.12.7841. [DOI] [PubMed] [Google Scholar]
- Lemmon G., Meiler J. Rosetta ligand docking with flexible XML protocols. Methods Mol. Biol. 2012;819:143–155. doi: 10.1007/978-1-61779-465-0_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner M., Carlson H. University of Michigan; Ann Arbor: 2020. APBS Plugin for PyMOL. [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S., Craig E.A. The heat-shock proteins. Annu. Rev. Genet. 1988;22:631–677. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- Liu Y., Steiniger S.C.J., Kim Y., Kaufmann G.F., Felding-Habermann B., Janda K.D. Mechanistic studies of a peptidic GRP78 ligand for cancer cell-specific drug delivery. Mol. Pharm. 2007;4:435–447. doi: 10.1021/mp060122j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xiao X., Wei X., Li J., Yang J., Tan H., Zhu J., Zhang Q., Wu J., Liu L. Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2. J. Med. Virol. 2020;92:595–601. doi: 10.1002/jmv.25726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2020) Maestro, Schrödinger, LLC, New York, NY.
- Manikyam H.K., Joshi S.K. Whole genome analysis and targeted drug discovery using computational methods and high throughput screening tools for emerged novel coronavirus (2019-nCoV) J. Pharmaceut. Drug Res. 2020;3:341–361. [PMC free article] [PubMed] [Google Scholar]
- Mehboob, R., Ahmad, F.J., Qayyum, A., Rana, M.A., Gilani, S.A., Tariq, M.A., Ali, G., Akram, S.J., and Akram, J.. (2020) Aprepitant as a combinant with Dexamethasone reduces the inflammation via Neurokinin 1 Receptor Antagonism in severe to critical Covid-19 patients and potentiates respiratory recovery: a novel therapeutic approach, medRxiv, 2020.2008.2001.20166678.
- Misra, U.K., Gonzalez-Gronow, M. Fau - Gawdi, G., Gawdi, G. Fau - Pizzo, S.V., and Pizzo, S.V. (2005) The role of MTJ-1 in cell surface translocation of GRP78, a receptor for alpha 2-macroglobulin-dependent signaling. [DOI] [PubMed]
- Misra U.K., Mowery Y., Kaczowka S., Pizzo S.V. Ligation of cancer cell surface GRP78 with antibodies directed against its COOH-terminal domain up-regulates p53 activity and promotes apoptosis. Mol. Cancer Ther. 2009;8:1350–1362. doi: 10.1158/1535-7163.MCT-08-0990. [DOI] [PubMed] [Google Scholar]
- Misra U.K., Pizzo S.V. Modulation of the unfolded protein response in prostate cancer cells by antibody-directed against the carboxyl-terminal domain of GRP78. Apoptosis. 2010;15:173–182. doi: 10.1007/s10495-009-0430-y. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nain M., Mukherjee S., Karmakar S.P., Paton A.W., Paton J.C., Abdin M.Z., Basu A., Kalia M., Vrati S. GRP78 Is an important host factor for Japanese encephalitis virus entry and replication in mammalian cells. J. Virol. 2017;91 doi: 10.1128/JVI.02274-16. e02274-02216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair P., Lu M., Petersen S., Ashkenazi A. Apoptosis initiation through the cell-extrinsic pathway. Methods Enzymol. 2014;544:99–128. doi: 10.1016/B978-0-12-417158-9.00005-4. [DOI] [PubMed] [Google Scholar]
- Nakao K., Murase A., Ohshiro H., Okumura T., Taniguchi K., Murata Y., Masuda M., Kato T., Okumura Y., Takada J. CJ-023,423, a novel, potent and selective prostaglandin EP4 receptor antagonist with antihyperalgesic properties. J. Pharmacol. Exp. Ther. 2007;322:686–694. doi: 10.1124/jpet.107.122010. [DOI] [PubMed] [Google Scholar]
- Naming the coronavirus disease (COVID-19) and the virus that causes it, World Health Organization, Technical Guidance.
- O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open babel: an open chemical toolbox. J. Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeira A., Sousa E., Koseler A., Sabirli R., Goren T., Turkcuer I., Kurt O., Pinto M.M., Vasconcelos M.H. Preliminary virtual screening studies to identify GRP78 inhibitors which may interfere with SARS-CoV-2 infection. Pharmaceuticals (Basel) 2020;13 doi: 10.3390/ph13060132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujhari S., Brustolin M., Macias V.M., Nissly R.H., Nomura M., Kuchipudi S.V., Rasgon J.L. Heat shock protein 70 (Hsp70) mediates Zika virus entry, replication, and egress from host cells. Emerg. Microbes Infect. 2019;8:8–16. doi: 10.1080/22221751.2018.1557988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid S.P., Shurtleff A.C., Costantino J.A., Tritsch S.R., Retterer C., Spurgers K.B., Bavari S. HSPA5 is an essential host factor for Ebola virus infection. Antiviral Res. 2014;109:171–174. doi: 10.1016/j.antiviral.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Robb C.T., Goepp M., Rossi A.G., Yao C. Non-steroidal anti-inflammatory drugs, prostaglandins, and COVID-19. Br. J. Pharmacol. 2020;177:4899–4920. doi: 10.1111/bph.15206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R., Nillegoda N.B., Mayer M.P., Bukau B. The Hsp70 chaperone network. Nat. Rev. Mol. Cell Biol. 2019;20:665–680. doi: 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- Royle J., Ramírez-Santana C., Akpunarlieva S., Donald C.L., Gestuveo R.J., Anaya J.M., Merits A., Burchmore R., Kohl A., Varjak M. Glucose-regulated protein 78 interacts with Zika virus envelope protein and contributes to a productive infection. Viruses. 2020;12 doi: 10.3390/v12050524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiq S., Owen E., Foster T., Knight K., Wang L., Pirmohamed M., Clark R.E., Pushpakom S. Nilotinib-induced metabolic dysfunction: insights from a translational study using in vitro adipocyte models and patient cohorts. Leukemia. 2019;33:1810–1814. doi: 10.1038/s41375-018-0337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2020-4: LigPrep, Schrödinger, LLC, New York, NY, 2020.
- Schrodinger, LLC. (2015) The PyMOL molecular graphics system, Version 1.8.
- Shah B., Modi P., Sagar S.R. In silico studies on therapeutic agents for COVID-19: drug repurposing approach. Life Sci. 2020;252 doi: 10.1016/j.lfs.2020.117652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.P., Kipar A., Cox H., Payne C., Vasiliou S., Quinn J.P. Induction of tachykinin production in airway epithelia in response to viral infection. PLoS One. 2008;3:e1673. doi: 10.1371/journal.pone.0001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvas S. Role of substance P neuropeptide in inflammation, wound healing, and tissue homeostasis. J. Immunol. 2017;199:1543–1552. doi: 10.4049/jimmunol.1601751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triantafilou K., Fradelizi D., Wilson K., Triantafilou M. GRP78, a coreceptor for coxsackievirus A9, interacts with major histocompatibility complex class I molecules which mediate virus internalization. J. Virol. 2002;76:633–643. doi: 10.1128/JVI.76.2.633-643.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-L., Lee A.S. In: Cell Surface GRP78, a New Paradigm in Signal Transduction Biology. Pizzo S.V., editor. Academic Press; 2018. Chapter 3 - cell surface GRP78: anchoring and translocation mechanisms and therapeutic potential in cancer; pp. 41–62. [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R., Zou X., Hoyle G.W. Tachykinin-1 receptor stimulates proinflammatory gene expression in lung epithelial cells through activation of NF-κB via a Gq-dependent pathway. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2007;292:L430–L437. doi: 10.1152/ajplung.00475.2005. [DOI] [PubMed] [Google Scholar]
- Yang J., Nune M., Zong Y., Zhou L., Liu Q. Close and allosteric opening of the polypeptide-binding site in a human Hsp70 chaperone BiP. Structure. 2015;23:2191–2203. doi: 10.1016/j.str.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zong Y., Su J., Li H., Zhu H., Columbus L., Zhou L., Liu Q. Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s. Nat. Commun. 2017;8:1201. doi: 10.1038/s41467-017-01310-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G., Lee A.S. Role of the unfolded protein response, GRP78 and GRP94 in organ homeostasis. J. Cell. Physiol. 2015;230:1413–1420. doi: 10.1002/jcp.24923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The docking structures are available upon request.