Fig. 2.
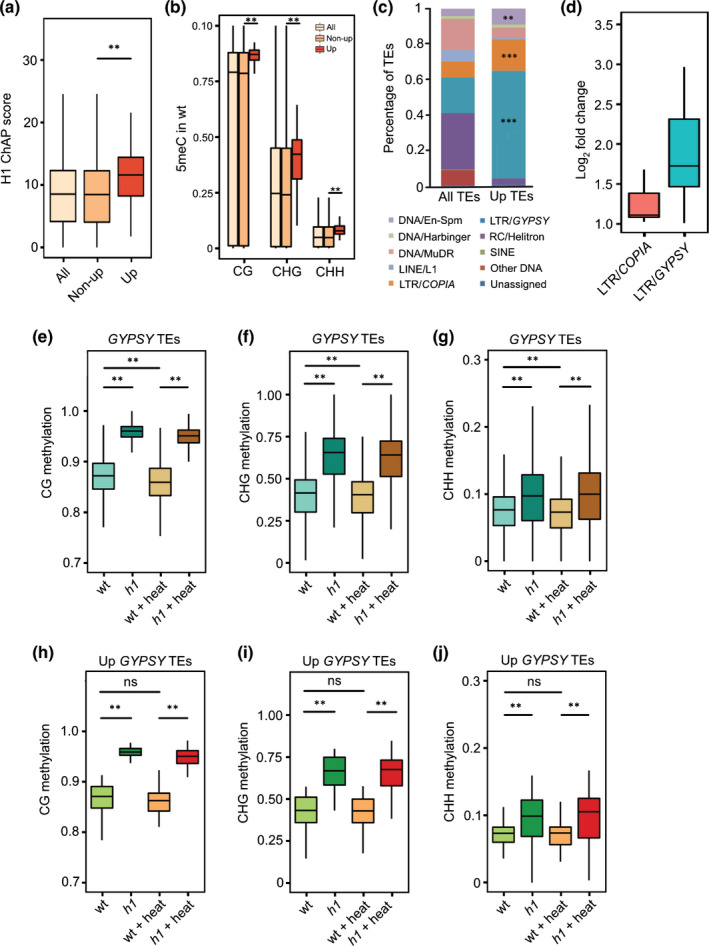
Histone H1‐mediated repression of GYPSY transposable elements (TEs) upon heat occurs independently of DNA methylation in Arabidopsis. Wild‐type (WT) H1 enrichment (a) on all, nonupregulated and upregulated TEs in h1 plus heat vs WT plus heat. **, P < 0.01 (Wilcoxon test). ChAP, chromatin affinity purification. H1 ChAP score was defined as the average ChAP coverage from start to end of the TE. Wild‐type DNA methylation level (CG, CHG and CHH, (b)) on all, nonupregulated and upregulated TEs in h1 plus heat vs WT plus heat. **, P < 0.01 (Wilcoxon test). Percentages of TEs are classified in different families (c). Up TEs refer to the upregulated TEs in h1 plus heat vs WT plus heat. **, P < 0.01; ***, P < 0.001 (hypergeometric test). Expression level (log2‐fold change, (d)) of upregulated long terminal repeat (LTR)/COPIA and LTR/GYPSY elements in h1 plus heat vs WT plus heat. Averaged CG (e), CHG (f) and CHH (g) methylation level on all GYPSY TEs in WT, h1, WT plus heat (WT + heat) and h1 plus heat (h1 + heat). Averaged CG (h), CHG (i) and CHH (j) methylation on upregulated GYPSY TEs in h1 plus heat vs WT plus heat in four different conditions of WT, h1, WT plus heat and h1 plus heat. **, P < 0.01; ns, not significant (Wilcoxon test). The lower and upper hinges of the boxplots correspond to the first and third quartiles of the data, the black lines within the boxes mark the median.
