Fig. 4.
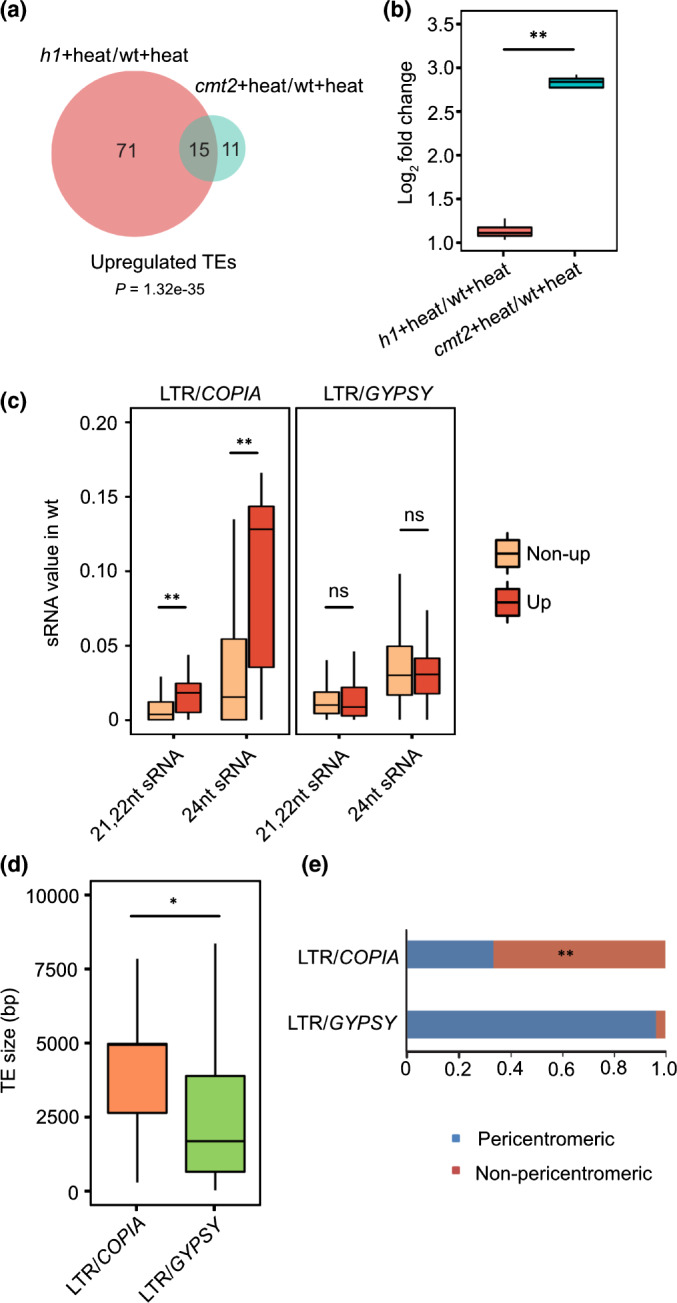
CHROMOMETHYLASE 2 (CMT2) is required for COPIA78/ONSEN transposable element (TE) repression under heat conditions in Arabidopsis. Venn diagram (a) shows upregulated TEs in h1 plus heat vs wild‐type (WT) plus heat (h1 + heat/WT + heat) and cmt2 plus heats wt plus heat (cmt2 + heat/WT + heat). Boxplot (b) shows the expression level of commonly upregulated COPIA78/ONSEN elements in h1 plus heat vs WT plus heat and cmt2 plus heat vs WT plus heat. **, P < 0.01 (Wilcoxon test). Boxplot (c) shows WT sRNA value on upregulated and nonupregulated long terminal repeat (LTR)/COPIA and LTR/GYPSY elements in h1 plus heat vs WT plus heat. **, P < 0.01; ns, not significant (Wilcoxon test). Boxplot (d) shows the size of upregulated LTR/COPIA TEs and LTR/GYPSY TEs in h1 plus heat vs WT plus heat. *, P < 0.05 (Wilcoxon test). The lower and upper hinges of the boxplots correspond to the first and third quartiles of the data, the black lines within the boxes mark the median. The percentages of upregulated LTR/COPIA and LTR/GYPSY‐type TEs in h1 plus heat vs WT plus heat are classified by location (e). Upregulated COPIA elements are significantly enriched in nonpericentromeric regions compared to all LTR/COPIA TEs. **, P < 0.01 (hypergeometric test).
