Abstract
Among various delivery systems for vaccine and drug delivery, cell-penetrating peptides (CPPs) have been known as a potent delivery system because of their capability to penetrate cell membranes and deliver some types of cargoes into cells. Several CPPs were found in the proteome of viruses such as Tat originated from human immunodeficiency virus-1 (HIV-1), and VP22 derived from herpes simplex virus-1 (HSV-1). In the current study, a wide-range of CPPs was identified in the proteome of SARS-CoV-2, a new member of coronaviruses family, using in silico analyses. These CPPs may play a main role for high penetration of virus into cells and infection of host. At first, we submitted the proteome of SARS-CoV-2 to CellPPD web server that resulted in a huge number of CPPs with ten residues in length. Afterward, we submitted the predicted CPPs to C2Pred web server for evaluation of the probability of each peptide. Then, the uptake efficiency of each peptide was investigated using CPPred-RF and MLCPP web servers. Next, the physicochemical properties of the predicted CPPs including net charge, theoretical isoelectric point (pI), amphipathicity, molecular weight, and water solubility were calculated using protparam and pepcalc tools. In addition, the probability of membrane binding potential and cellular localization of each CPP were estimated by Boman index using APD3 web server, D factor, and TMHMM web server. On the other hand, the immunogenicity, toxicity, allergenicity, hemolytic potency, and half-life of CPPs were predicted using various web servers. Finally, the tertiary structure and the helical wheel projection of some CPPs were predicted by PEP-FOLD3 and Heliquest web servers, respectively. These CPPs were divided into: a) CPP containing tumor homing motif (RGD) and/or tumor penetrating motif (RXXR); b) CPP with the highest Boman index; c) CPP with high half-life (~100 hour) in mammalian cells, and d) CPP with +5.00 net charge. Based on the results, we found a large number of novel CPPs with various features. Some of these CPPs possess tumor-specific motifs which can be evaluated in cancer therapy. Furthermore, the novel and potent CPPs derived from SARS-CoV-2 may be used alone or conjugated to some sequences such as nuclear localization sequence (NLS) for vaccine and drug delivery.
Introduction
Therapeutic and preventive vaccines are promising approaches to solve health issues globally [1]. Although there are several vaccines for saving millions of lives till now such as vaccines against rubella, mumps, varicella, rotavirus, human papillomavirus (HPV) and hepatitis B virus (HBV), it is required to develop effective vaccines against other pathogens which are incurable and unprotectable [2,3]. In this line, development of effective and novel delivery systems is vital for delivery of vaccine components into cells. In general, delivery systems can be used to transfer different biomolecules into cells including nanoparticles [4], polymers [5], chitosan [6], liposome [7], physical tools [8], and cell penetrating peptides (CPPs) [9,10]. The current focus of developing a novel delivery system has moved to peptide-based delivery systems known as CPPs [11]. CPPs contain 5–50 amino acids in length which can enter cell membranes efficiently and deliver a wide range of cargoes including peptides, proteins, nanoparticles and nucleic acids into cells [12,13]. After discovery of the first CPP, Tat peptide (originated from human immunodeficiency virus type-1 (HIV-1) trans-activating regulatory (Tat) protein), a rapid growth of new CPPs has occurred [14]. The CPPs are natural (e.g., CyLoP-1) or synthetic (e.g., oligoarginine) peptides. These short peptides are heterogeneous in sequence and structure, and can be delivered through endocytosis or direct penetration [1,10,15–17]. The mechanism of internalization depends on diverse factors such as CPP sequence, cell type, CPP concentration, temperature, incubation time, and type of cargo [18]. Up to now, a large number of CPPs have been recognized but some of them showed low uptake [19]. The studies demonstrated that prediction of CPPs by bioinformatics tools prior to lab-based experiments could save time and money [20]. For instance, machine-learning-based algorithms permit users to predict CPPs from large sequence data/ proteome. In prediction methods, machine learning models utilize various algorithms including neural network (NN) [21,22], kernel extreme learning machine [23,24], random forest (RF) [25], and support vector machine (SVM) [26,27].
In December 2019, a new member of the coronavirus family was found which firstly named as 2019-nCoV. Then, on February 11, 2020, its name was changed to Coronavirus Disease-2019 (COVID-19) or severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [28]. SARS-CoV-2 is an enveloped positive single-strand RNA virus that has 10 open reading frames (ORFs). ORF1ab is about 66.66% of virus genome which encodes two large polypeptides such as pp1a and pp1ab. Meanwhile, the ORF2-10 is about 33.33% of virus genome. SARS-CoV-2 genome encodes 28 proteins which are classified into three various classes such as structural proteins, non-structural proteins (nsp), and accessory proteins. The structural proteins are spike (S), nucleoprotein (N), membrane (M), and envelope (E) proteins that form the virus particles. In addition, the non-structural proteins (e.g., nsp1-nsp16) are generated only during the translation of virus RNA in the infected host cell. The accessory proteins possess crucial functions in the assembly, virulence and pathogenesis of the virus [28,29].
Previously, several CPPs were derived from viruses such as Tat (from HIV-1 transcriptional activator protein), C105Y (from HIV-1 glycoprotein 41), MPG (from HIV-1 glycoprotein 41 conjugated to nuclear localization sequence (NLS) from simian virus 40 (SV40)), Pep-1 (from HIV-1 reverse transcriptase conjugated to SV40 NLS), pepR and pepM (originated from Dengue virus), VP22 (originated from Herpes simplex virus (HSV)-1) [14,30–34]. Up to now, no complete report has been available on CPPs derived from total proteins of MERS-CoV, SARS-CoV and SARS-CoV-2. Few studies indicated that some peptides of SARS-CoV spike glycoprotein are responsible for membrane fusion or membrane binding activity. For example, the upstream region of the heptad repeat1 (HR1) (residues 892–972) in S2 domain of SARS-CoV spike glycoprotein was involved in membrane fusion. Moreover, some scientists have recognized membrane binding peptides and membrane fusogenic peptides or potential fusion peptides from the upstream region of HR1 (residues 758–890) [35]. Indeed, an efficient membrane fusion mechanism between host cell and SARS-CoV-2 can be responsible for virus infection. Sequence comparison of S protein domains between SARS-CoV-2 and SARS-CoV-1 showed high level of conservation for both S1 and S2 domains. However, variation in the fusogenic regions of S2 domain was observed between SARS-CoV-2 and SARS-CoV-1 [36–38]. Hence, due to high potency of SARS-CoV-2 to spread and infect people, we decided to investigate new and potent CPPs in the proteome of this newly isolated virus using in silico approaches.
Materials and methods
Study design
The current study has several main steps to find and characterize novel and potent CPPs as a vaccine and drug delivery system. The flowchart of overall prediction and analysis procedure was illustrated in S1 Fig.
Identification of potential SARS-CoV-2-derived CPPs
Cell penetrating or non-cell penetrating peptides (CPP or non-CPP) could be predicted in the proteome of SARS-CoV-2 using bioinformatics approaches. Hence, to explore novel CPPs, our reference sequence was Wuhan-Hu-1 with GenBank accession number MN908947.3. This strain was isolated from a patient in Wuhan, china. The phylogenetic analysis of whole viral genome contain 29,903 nucleotides that has 89.1% nucleotide similarity to a group of SARS-like coronaviruses (genus Betacoronavirus, subgenus Sarbecovirus) which formerly had been recognized in bats [39].
At first, CellPPD web server (https://webs.iiitd.edu.in/raghava/cellppd/index.html) was applied to determine the cell penetrating peptides. CellPPD is a support vector machine (SVM)-based web server [40,41]. To utilize this web server, the sequences of Spike (S) protein (GenBank ID: QHD43416.1), Membrane (M) glycoprotein (GenBank ID: QHD43419.1), Nucleocapsid (N) phosphoprotein (GenBank ID: QHD43423.2), Envelope (E) protein (GenBank ID: QHD43418.1), Orf1ab polyprotein (GenBank ID: QHD43415.1), ORF3a protein (GenBank ID: QHD43417.1), ORF6 protein (GenBank ID: QHD43420.1), ORF7a protein (GenBank ID: QHD43421.1), ORF8 protein (GenBank ID: QHD43422.1), and ORF10 protein (GenBank ID: QHI42199.1) were submitted to protein scanning tool with default threshold of the SVM-based prediction method (SVM threshold was set at 0.0). Moreover, Tang and colleagues [42] developed a method with an overall prediction accuracy of 83.6%; hence, they established C2Pred web server (http://lin-group.cn/server/C2Pred) to investigate the CPP probability of each peptide.
Uptake efficiency analysis of the identified CPPs
In next step, to evaluate the uptake efficiency of the identified CPPs from previous step, two web servers including CPPred-RF (http://server.malab.cn/CPPred-RF/), and MLCPP (http://www.thegleelab.org/MLCPP/) were used. For this purpose, all of the detected CPPs using CellPPD web server were submitted to these web servers. CPPred-RF is a sequence-based predictor for identifying CPPs and their uptake efficiency. In addition, CPPred-RF built a two-layer prediction framework according to the random forest (RF) algorithm [43]. Manavalan et al. established a two-layer prediction framework termed as machine-learning-based prediction of cell-penetrating peptide (MLCPPs). The first-layer predicts that a submitted peptide is categorized as a CPP or non-CPP. Meanwhile, the second-layer predicts the uptake efficiency of the predicted CPPs [44].
Peptides property calculation
It is crucial to compute the physicochemical properties of peptides for predicting and designing novel and potent CPPs. Therefore, to achieve this aim, we calculated various physicochemical features of CPPs such as net charge, theoretical isoelectric point (pI), amphipathicity, molecular weight (MW), water solubility, hydrophobicity (H), hydrophobicity ratio, and polar-, non-polar-, uncharged- and charged residues. To calculate net charge, theoretical pI, and amphipathicity, CellPPD web server (https://webs.iiitd.edu.in/raghava/cellppd/index.html) was utilized. In addition, protparam tool (https://web.expasy.org/protparam/) was used to compute molecular weight of CPPs. Furthermore, to obtain the water solubility of peptides, Peptide property calculator (PepCalc) (https://pepcalc.com/) was applied. Also, hydrophobicity (H), and polar-, non-polar-, uncharged- and charged residues were estimated using Heliquest web server (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py).
Evaluation of membrane-binding ability of CPPs
In order to investigate the potential of binding peptides to membrane, two different methods were utilized. At first, we evaluated the Boman index or protein-binding potential using APD3 web server (http://aps.unmc.edu/AP/prediction/prediction_main.php). The Boman index is the sum of solubility values for all presented amino acids in a peptide sequence and illustrates the potential of a peptide for binding to the membrane or other proteins [45]. Secondly, to evaluate the membrane-binding potential of each peptide, the discrimination factor (D) was calculated [46]. For this purpose, we used Heliquest web server (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py) to obtain hydrophobic moment (μH). After determination of hydrophobic moment and also net charge (Z), the D factor was calculated according to the following equation: D = 0.944(<μH>) + 0.33(Z).
In addition, TMHMM web server (http://www.cbs.dtu.dk/services/TMHMM/) was utilized to investigate the cellular localization of CPPs [47]. This web server analyzes the probability of binding a peptide to the bacterial cell membrane (BCM) which possesses negative charge.
Assessment of the immunogenicity
Immunogenicity of the CPPs is one of their disadvantages. It was confirmed that peptides could induce immunologic responses in vivo, resulting in allergic reactions. The existence of peptides in body can stimulate the generation of antibodies which may neutralize therapeutic effects and reduce their efficacy [48, 49]. Hence, to assess the immunogenicity of CPPs, each peptide was submitted to IEDB Immunogenicity Predictor (http://tools.iedb.org/immunogenicity/) [50].
Determination of toxicity and allergenicity
To investigate the toxicity and allergenicity of CPPs, each peptide was submitted to ToxinPred web server (https://webs.iiitd.edu.in/raghava/toxinpred/algo.php), and AllerTop (https://www.ddg-pharmfac.net/AllerTOP/) and AllergenFP (http://ddg-pharmfac.net/AllergenFP/) web servers, respectively [51–53].
Estimation of hemolytic potency and half-life
The hemolytic property of peptides was predicted by HemoPI using SVM-based method (https://webs.iiitd.edu.in/raghava/hemopi/design.php). Furthermore, the half-life in E.coli and in mammalian cell was calculated using ProtLifePred web server based on N-end rule (http://protein-n-end-rule.leadhoster.com/) [54].
Prediction of structure
Three dimensional (3D) structure of some predicted CPPs was analyzed by de novo peptide structure prediction server (PEP-FOLD3) (https://bioserv.rpbs.univ-paris-diderot.fr/services/PEP-FOLD3/). PEP-FOLD3 is a de novo method that predicts peptide structures using amino acid sequences. This approach determines the conformation of four consecutive amino acid residues according to structural alphabet (SA) letters [55]. Additionally, the helical wheel diagram of CPPs was defined by Schiffer Edmundson wheel modelling using Heliquest web server (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py) [46].
Results
Identification of potential SARS-CoV-2-derived CPPs
To obtain cell penetrating peptides in the proteome of SARS-CoV-2, the sequences of S protein, M glycoprotein, N phosphoprotein, E protein, Orf1ab polyprotein, and ORF3a, ORF6, ORF7a, ORF8 and ORF10 proteins were submitted to protein scanning tools of CellPPD web server. Then, we applied C2Pred web server to achieve the CPP probability of peptides. All of the detected CPPs, and their SVM scores and probability scores were listed in Table 1. No CPP was found in E protein, and only one CPP was identified in ORF6. Meanwhile, Orf1ab had the most CPPs in its proteome. C2pred web server identifies peptides lower than 0.5 as non-CPPs, and peptides greater than 0.5 as CPPs. Although, some peptides were predicted as CPPs by CellPPD, but C2Pred detected them as non-CPPs. For instance, DMSKFPLKLR peptide derived from Orf1ab polyprotein was predicted as CPP by CellPPD with SVM score of 0.11, while C2Pred determined this peptide as non-CPP with score 0.167688.
Table 1. Predicted CPPs and their uptake efficiency using various web servers.
| Epitope | CellPPD (SVM score) | CPP Probability score by C2Pred* | Uptake efficiency by MLCPP | Uptake efficiency by CPPred-RF |
|---|---|---|---|---|
| S protein | ||||
| NLTTRTQLPP | 0.03 | 0.549136 | Low | High |
| RFQTLLALHR | 0.11 | 0.924753 | Low | High |
| YLQPRTFLLK | 0.05 | 0.745663 | Low | High |
| SVYAWNRKRI | 0.12 | 0.542719 | Low | High |
| YAWNRKRISN | 0.21 | 0.581372 | Low | High |
| AWNRKRISNC | 0.10 | 0.581372 | Low | High |
| WNRKRISNCV | 0.07 | 0.422121 | Low | High |
| RQIAPGQTGK | 0.05 | 0.480859 | Low | High |
| YNYLYRLFRK | 0.10 | 0.434133 | High | High |
| YLYRLFRKSN | 0.17 | 0.323746 | High | High |
| YRLFRKSNLK | 0.18 | 0.651294 | Low | High |
| RLFRKSNLKP | 0.18 | 0.633622 | Low | High |
| RKSNLKPFER | 0.14 | 0.800554 | Low | High |
| KKSTNLVKNK | 0.33 | 0.588285 | Low | High |
| KSTNLVKNKC | 0.06 | 0.527724 | Low | High |
| HADQLTPTWR | 0.00 | 0.545188 | Low | Non-CPP |
| YQTQTNSPRR | 0.11 | 0.149319 | Low | High |
| TQTNSPRRAR | 0.11 | 0.143146 | Low | High |
| TNSPRRARSV | 0.16 | 0.142480 | Low | High |
| NSPRRARSVA | 0.14 | 0.108221 | Low | High |
| PRRARSVASQ | 0.17 | 0.236319 | Low | High |
| KQIYKTPPIK | 0.46 | 0.944703 | Low | High |
| SQILPDPSKP | 0.03 | 0.906582 | Low | High |
| RLITGRLQSL | 0.21 | 0.663600 | Low | High |
| M protein | ||||
| NRNRFLYIIK | 0.02 | 0.220554 | Low | High |
| RNRFLYIIKL | 0.18 | 0.114117 | Low | High |
| YIIKLIFLWL | 0.01 | 0.669640 | Low | High |
| KLIFLWLLWP | 0.31 | 0.709344 | Low | High |
| FIASFRLFAR | 0.10 | 0.421173 | Low | High |
| ASFRLFARTR | 0.20 | 0.594022 | Low | High |
| SFRLFARTRS | 0.06 | 0.553645 | Low | High |
| FRLFARTRSM | 0.21 | 0.385465 | Low | High |
| RLFARTRSMW | 0.28 | 0.383433 | Low | High |
| FARTRSMWSF | 0.04 | 0.463913 | Low | High |
| HGTILTRPLL | 0.03 | 0.430916 | Low | High |
| GAVILRGHLR | 0.13 | 0.223997 | High | High |
| RIAGHHLGRC | 0.02 | 0.243928 | Low | High |
| YSRYRIGNYK | 0.05 | 0.897477 | Low | High |
| N protein | ||||
| PQNQRNAPRI | 0.09 | 0.413323 | Low | High |
| ERSGARSKQR | 0.06 | 0.261718 | Low | High |
| RSGARSKQRR | 0.54 | 0.292041 | Low | High |
| SGARSKQRRP | 0.25 | 0.300551 | Low | High |
| GARSKQRRPQ | 0.40 | 0.250417 | Low | High |
| ARSKQRRPQG | 0.17 | 0.298689 | Low | High |
| RSKQRRPQGL | 0.36 | 0.192042 | Low | High |
| SKQRRPQGLP | 0.06 | 0.164110 | Low | High |
| KQRRPQGLPN | 0.18 | 0.206509 | Low | High |
| RRPQGLPNNT | 0.12 | 0.216779 | Low | High |
| QIGYYRRATR | 0.09 | 0.531651 | Low | High |
| IGYYRRATRR | 0.23 | 0.604819 | Low | High |
| GYYRRATRRI | 0.29 | 0.965070 | Low | High |
| YYRRATRRIR | 0.58 | 0.952696 | Low | High |
| YRRATRRIRG | 0.59 | 0.935039 | Low | High |
| RRATRRIRGG | 0.56 | 0.939794 | Low | High |
| RATRRIRGGD | 0.33 | 0.924855 | Low | High |
| TRRIRGGDGK | 0.10 | 0.369598 | Low | High |
| RIRGGDGKMK | 0.11 | 0.250269 | Low | High |
| GKMKDLSPRW | 0.02 | 0.735642 | Low | High |
| SQASSRSSSR | 0.05 | 0.677066 | Low | High |
| ASSRSSSRSR | 0.17 | 0.304528 | Low | High |
| SSRSSSRSRN | 0.09 | 0.304528 | Low | High |
| SRSSSRSRNS | 0.14 | 0.297448 | Low | High |
| RSSSRSRNSS | 0.12 | 0.432735 | Low | High |
| SSSRSRNSSR | 0.13 | 0.315140 | Low | High |
| SSRSRNSSRN | 0.10 | 0.315140 | Low | High |
| RSRNSSRNST | 0.04 | 0.359073 | Low | High |
| GSSRGTSPAR | 0.06 | 0.716403 | Low | High |
| AALALLLLDR | 0.14 | 0.898706 | Low | High |
| ALALLLLDRL | 0.05 | 0.895190 | Low | High |
| KKSAAEASKK | 0.33 | 0.730535 | Low | High |
| KSAAEASKKP | 0.13 | 0.669679 | Low | High |
| SAAEASKKPR | 0.06 | 0.669679 | Low | High |
| AAEASKKPRQ | 0.10 | 0.963042 | Low | High |
| AEASKKPRQK | 0.15 | 0.96509 | Low | High |
| EASKKPRQKR | 0.34 | 0.988105 | Low | High |
| ASKKPRQKRT | 0.28 | 0.947786 | Low | High |
| SKKPRQKRTA | 0.32 | 0.947786 | Low | High |
| KKPRQKRTAT | 0.47 | 0.947786 | Low | High |
| KPRQKRTATK | 0.42 | 0.941370 | Low | High |
| PRQKRTATKA | 0.23 | 0.936878 | Low | High |
| RQKRTATKAY | 0.13 | 0.936878 | Low | High |
| RQGTDYKHWP | 0.02 | 0.249058 | Low | High |
| FPPTEPKKDK | 0.04 | 0.937092 | Low | Non-CPP |
| PPTEPKKDKK | 0.12 | 0.954110 | Low | High |
| PTEPKKDKKK | 0.20 | 0.965728 | Low | High |
| TEPKKDKKKK | 0.51 | 0.972132 | Low | High |
| EPKKDKKKKA | 0.35 | 0.969009 | Low | High |
| PKKDKKKKAD | 0.28 | 0.736106 | Low | High |
| KKDKKKKADE | 0.26 | 0.275127 | Low | High |
| TQALPQRQKK | 0.11 | 0.378326 | Low | High |
| ALPQRQKKQQ | 0.17 | 0.715403 | Low | High |
| ORF3a | ||||
| SASKIITLKK | 0.07 | 0.666607 | Low | High |
| ASKIITLKKR | 0.19 | 0.975854 | Low | High |
| SKIITLKKRW | 0.31 | 0.977206 | Low | High |
| KIITLKKRWQ | 0.56 | 0.977206 | Low | High |
| IITLKKRWQL | 0.04 | 0.981679 | Low | High |
| KKRWQLALSK | 0.47 | 0.908996 | Low | High |
| KRWQLALSKG | 0.31 | 0.711236 | Low | High |
| VRIIMRLWLC | 0.11 | 0.512905 | Low | High |
| RIIMRLWLCW | 0.25 | 0.512905 | Low | High |
| IIMRLWLCWK | 0.14 | 0.584110 | Low | High |
| IMRLWLCWKC | 0.06 | 0.584110 | High | High |
| MRLWLCWKCR | 0.17 | 0.584110 | High | High |
| RLWLCWKCRS | 0.22 | 0.578205 | High | High |
| LWLCWKCRSK | 0.28 | 0.516957 | High | High |
| WLCWKCRSKN | 0.12 | 0.456487 | High | High |
| LCWKCRSKNP | 0.16 | 0.456487 | High | High |
| CWKCRSKNPL | 0.14 | 0.815386 | High | High |
| KCRSKNPLLY | 0.10 | 0.172543 | Low | High |
| Orf1ab | ||||
| IKRSDARTAP | 0.00 | 0.611515 | Low | High |
| KRSDARTAPH | 0.08 | 0.611515 | Low | High |
| PVAYRKVLLR | 0.05 | 0.395891 | High | High |
| VAYRKVLLRK | 0.07 | 0.461402 | High | High |
| AYRKVLLRKN | 0.13 | 0.873577 | Low | High |
| YRKVLLRKNG | 0.05 | 0.873577 | Low | High |
| RKVLLRKNGN | 0.14 | 0.892119 | Low | High |
| KVLLRKNGNK | 0.01 | 0.921012 | Low | High |
| FEIKLAKKFD | 0.09 | 0.877125 | Low | High |
| KTIQPRVEKK | 0.39 | 0.961264 | Low | High |
| TIQPRVEKKK | 0.13 | 0.954110 | Low | High |
| IQPRVEKKKL | 0.13 | 0.953833 | Low | High |
| SGLKTILRKG | 0.02 | 0.624757 | Low | High |
| LKTILRKGGR | 0.24 | 0.808425 | Low | High |
| KTILRKGGRT | 0.15 | 0.805867 | Low | High |
| GNFKVTKGKA | 0.03 | 0.385736 | Low | High |
| FKVTKGKAKK | 0.35 | 0.445305 | Low | High |
| KVTKGKAKKG | 0.26 | 0.339767 | Low | High |
| KGKAKKGAWN | 0.07 | 0.184903 | Low | High |
| GGAKLKALNL | 0.04 | 0.430832 | Low | High |
| SKGLYRKCVK | 0.09 | 0.145729 | Low | High |
| KGLYRKCVKS | 0.07 | 0.145729 | Low | High |
| GLYRKCVKSR | 0.34 | 0.274849 | Low | High |
| GLLMPLKAPK | 0.09 | 0.427961 | Low | High |
| QRKQDDKKIK | 0.15 | 0.861718 | Low | High |
| RKQDDKKIKA | 0.17 | 0.976533 | Low | High |
| KQDDKKIKAC | 0.07 | 0.318404 | Low | High |
| DITFLKKDAP | 0.03 | 0.682254 | Low | Non-CPP |
| MLAKALRKVP | 0.28 | 0.671110 | High | High |
| LAKALRKVPT | 0.08 | 0.671110 | Low | High |
| EAKTVLKKCK | 0.13 | 0.717688 | Low | High |
| AKTVLKKCKS | 0.13 | 0.717688 | Low | High |
| KTVLKKCKSA | 0.22 | 0.546244 | Low | High |
| KSAFYILPSI | 0.00 | 0.108440 | Low | High |
| KAIVSTIQRK | 0.08 | 0.204524 | Low | High |
| STIQRKYKGI | 0.07 | 0.433511 | Low | High |
| TIQRKYKGIK | 0.13 | 0.433511 | Low | High |
| IQRKYKGIKI | 0.08 | 0.433511 | Low | High |
| GARFYFYTSK | 0.04 | 0.178363 | Low | High |
| ARYMRSLKVP | 0.06 | 0.471501 | Low | High |
| GIEFLKRGDK | 0.08 | 0.224238 | Low | High |
| DNLKTLLSLR | 0.03 | 0.361469 | High | Non-CPP |
| YMSALNHTKK | 0.05 | 0.330689 | Low | High |
| SALNHTKKWK | 0.12 | 0.709364 | Low | High |
| ALNHTKKWKY | 0.10 | 0.888647 | Low | High |
| LNHTKKWKYP | 0.14 | 0.913973 | Low | High |
| NHTKKWKYPQ | 0.10 | 0.678789 | Low | High |
| HTKKWKYPQV | 0.04 | 0.303493 | Low | Non-CPP |
| KKPASRELKV | 0.14 | 0.769021 | Low | High |
| KPASRELKVT | 0.07 | 0.743805 | Low | High |
| YTPSFKKGAK | 0.08 | 0.164140 | Low | High |
| PSFKKGAKLL | 0.00 | 0.210028 | Low | High |
| FKKGAKLLHK | 0.16 | 0.308486 | Low | High |
| KKGAKLLHKP | 0.48 | 0.308486 | Low | High |
| KGAKLLHKPI | 0.32 | 0.261312 | Low | High |
| WCIRCLWSTK | 0.12 | 0.727461 | High | High |
| CIRCLWSTKP | 0.20 | 0.727461 | Low | High |
| ANYAKPFLNK | 0.01 | 0.474322 | Low | High |
| TNIVTRCLNR | 0.01 | 0.309069 | Low | High |
| CTFTRSTNSR | 0.02 | 0.513802 | Low | High |
| TCMMCYKRNR | 0.01 | 0.331608 | High | High |
| MCYKRNRATR | 0.25 | 0.936560 | Low | High |
| CYKRNRATRV | 0.03 | 0.936560 | Low | High |
| YKRNRATRVE | 0.00 | 0.936560 | Low | High |
| KRNRATRVEC | 0.18 | 0.532289 | Low | High |
| RNRATRVECT | 0.06 | 0.325111 | Low | High |
| RDLSLQFKRP | 0.17 | 0.903779 | Low | High |
| SLQFKRPINP | 0.14 | 0.353192 | Low | High |
| HNIALIWNVK | 0.01 | 0.377627 | Low | High |
| LSEQLRKQIR | 0.04 | 0.269638 | Low | High |
| QLRKQIRSAA | 0.03 | 0.615417 | Low | High |
| LRKQIRSAAK | 0.02 | 0.718721 | Low | High |
| RKQIRSAAKK | 0.30 | 0.751730 | Low | High |
| KQIRSAAKKN | 0.38 | 0.729912 | Low | High |
| QIRSAAKKNN | 0.11 | 0.729912 | Low | High |
| AAKKNNLPFK | 0.09 | 0.933084 | Low | High |
| KKNNLPFKLT | 0.04 | 0.938618 | Low | High |
| NNWLKQLIKV | 0.01 | 0.793676 | High | High |
| LAYYFMRFRR | 0.04 | 0.301382 | Low | High |
| AYYFMRFRRA | 0.05 | 0.289065 | Low | High |
| YYFMRFRRAF | 0.08 | 0.182381 | High | High |
| FMRFRRAFGE | 0.05 | 0.167797 | High | High |
| MRFRRAFGEY | 0.05 | 0.167797 | High | High |
| KEMYLKLRSD | 0.02 | 0.354324 | Low | High |
| YNRYLALYNK | 0.03 | 0.241742 | High | High |
| RYLALYNKYK | 0.03 | 0.232393 | High | High |
| FRKMAFPSGK | 0.03 | 0.318037 | Low | High |
| TANPKTPKYK | 0.15 | 0.944703 | Low | High |
| ANPKTPKYKF | 0.03 | 0.944703 | Low | High |
| PKTPKYKFVR | 0.12 | 0.944703 | Low | High |
| KTPKYKFVRI | 0.10 | 0.917400 | Low | High |
| RWFLNRFTTT | 0.02 | 0.601321 | Low | High |
| FQSAVKRTIK | 0.08 | 0.586710 | Low | High |
| SEVVLKKLKK | 0.24 | 0.204172 | Low | High |
| VVLKKLKKSL | 0.09 | 0.270145 | Low | High |
| VLKKLKKSLN | 0.12 | 0.910975 | Low | High |
| KKLKKSLNVA | 0.15 | 0.190690 | Low | High |
| DAAMQRKLEK | 0.00 | 0.372134 | Low | High |
| AAMQRKLEKM | 0.01 | 0.372134 | Low | High |
| MQRKLEKMAD | 0.16 | 0.525121 | Low | High |
| YKQARSEDKR | 0.02 | 0.246035 | Low | High |
| KQARSEDKRA | 0.13 | 0.242658 | Low | High |
| QARSEDKRAK | 0.05 | 0.242658 | Low | High |
| MLFTMLRKLD | 0.05 | 0.894360 | Low | Non-CPP |
| QDLKWARFPK | 0.02 | 0.860963 | Low | High |
| DLKWARFPKS | 0.08 | 0.860963 | Low | Non-CPP |
| LKWARFPKSD | 0.22 | 0.397145 | Low | High |
| KGFCDLKGKY | 0.06 | 0.207285 | Low | High |
| GVSAARLTPC | 0.03 | 0.277471 | Low | High |
| GFAKFLKTNC | 0.01 | 0.119116 | Low | Non-CPP |
| KTNCCRFQEK | 0.08 | 0.481756 | Low | High |
| PHISRQRLTK | 0.28 | 0.636709 | Low | High |
| HISRQRLTKY | 0.24 | 0.636709 | Low | High |
| ISRQRLTKYT | 0.11 | 0.636709 | Low | High |
| SRQRLTKYTM | 0.01 | 0.835148 | Low | High |
| RQRLTKYTMA | 0.11 | 0.878279 | Low | High |
| GERVRQALLK | 0.09 | 0.849631 | Low | High |
| RVRQALLKTV | 0.09 | 0.932330 | High | High |
| KPYIKWDLLK | 0.01 | 0.876722 | Low | High |
| RLKLFDRYFK | 0.05 | 0.261079 | Low | High |
| KLFDRYFKYW | 0.04 | 0.219406 | High | Non-CPP |
| FPFNKWGKAR | 0.02 | 0.889091 | Low | High |
| KWGKARLYYD | 0.10 | 0.289722 | Low | High |
| YAISAKNRAR | 0.03 | 0.154021 | Low | High |
| AISAKNRART | 0.05 | 0.241258 | Low | High |
| KNRARTVAGV | 0.12 | 0.254056 | Low | High |
| NRQFHQKLLK | 0.14 | 0.872914 | Low | High |
| RQFHQKLLKS | 0.06 | 0.938861 | Low | High |
| RIMASLVLAR | 0.05 | 0.515705 | High | High |
| RNLQHRLYEC | 0.08 | 0.165926 | Low | Non-CPP |
| RLYECLYRNR | 0.15 | 0.219346 | Low | High |
| SLRCGACIRR | 0.10 | 0.297578 | High | High |
| RCGACIRRPF | 0.15 | 0.288701 | High | High |
| CGACIRRPFL | 0.06 | 0.282884 | High | High |
| GACIRRPFLC | 0.19 | 0.252159 | High | High |
| ACIRRPFLCC | 0.11 | 0.249754 | High | High |
| CIRRPFLCCK | 0.31 | 0.306758 | High | High |
| IRRPFLCCKC | 0.11 | 0.355792 | High | High |
| RRPFLCCKCC | 0.18 | 0.274986 | High | High |
| MSYYCKSHKP | 0.01 | 0.306949 | Low | High |
| ANTCTERLKL | 0.02 | 0.144577 | Low | High |
| SWEVGKPRPP | 0.02 | 0.577246 | Low | High |
| VGKPRPPLNR | 0.11 | 0.233226 | Low | High |
| GKPRPPLNRN | 0.16 | 0.742499 | Low | High |
| KALKYLPIDK | 0.20 | 0.613457 | Low | High |
| DKCSRIIPAR | 0.01 | 0.588037 | Low | High |
| KCSRIIPARA | 0.05 | 0.577426 | Low | High |
| CSRIIPARAR | 0.07 | 0.565072 | Low | High |
| SRIIPARARV | 0.11 | 0.783032 | Low | High |
| RIIPARARVE | 0.17 | 0.441924 | Low | High |
| SVVNARLRAK | 0.04 | 0.107521 | Low | High |
| VVNARLRAKH | 0.10 | 0.101852 | Low | High |
| VNARLRAKHY | 0.08 | 0.351672 | Low | High |
| NARLRAKHYV | 0.13 | 0.942607 | Low | High |
| PAPRTLLTKG | 0.10 | 0.701705 | Low | High |
| APRTLLTKGT | 0.02 | 0.414738 | Low | High |
| FNSVCRLMKT | 0.01 | 0.084142 | Low | Non-CPP |
| FLGTCRRCPA | 0.09 | 0.153800 | High | High |
| DNKLKAHKDK | 0.05 | 0.454919 | Low | High |
| KLKAHKDKSA | 0.22 | 0.624144 | Low | High |
| FLTRNPAWRK | 0.17 | 0.889087 | Low | High |
| RNPAWRKAVF | 0.18 | 0.548965 | Low | High |
| GIPKDMTYRR | 0.06 | 0.265085 | Low | High |
| DMTYRRLISM | 0.10 | 0.234218 | Low | Non-CPP |
| GNPKAIKCVP | 0.05 | 0.253610 | Low | High |
| WNTFTRLQSL | 0.06 | 0.961510 | Low | Non-CPP |
| ELWAKRNIKP | 0.06 | 0.982818 | Low | High |
| LWAKRNIKPV | 0.01 | 0.982818 | Low | High |
| WAKRNIKPVP | 0.17 | 0.862586 | Low | High |
| RNIKPVPEVK | 0.05 | 0.642094 | Low | High |
| LLIGLAKRFK | 0.19 | 0.367884 | High | High |
| GLAKRFKESP | 0.18 | 0.908642 | Low | High |
| KMQRMLLEKC | 0.17 | 0.500000 | High | High |
| VLRQWLPTGT | 0.00 | 0.407571 | Low | High |
| DMSKFPLKLR | 0.11 | 0.167688 | High | High |
| MSKFPLKLRG | 0.02 | 0.319169 | Low | High |
| SKFPLKLRGT | 0.09 | 0.463853 | Low | High |
| KFPLKLRGTA | 0.01 | 0.463853 | Low | High |
| MILSLLSKGR | 0.02 | 0.290394 | Low | High |
| LLSKGRLIIR | 0.08 | 0.570179 | High | High |
| GRLIIRENNR | 0.03 | 0.864554 | Low | High |
| RLIIRENNRV | 0.03 | 0.930275 | Low | High |
| ORF6 | ||||
| LIIKNLSKSL | 0.03 | 0.692987 | Low | High |
| ORF7a | ||||
| HVYQLRARSV | 0.00 | 0.333973 | Low | High |
| QLRARSVSPK | 0.18 | 0.788277 | Low | High |
| RARSVSPKLF | 0.06 | 0.540759 | Low | High |
| RSVSPKLFIR | 0.03 | 0.387766 | Low | High |
| ITLCFTLKRK | 0.00 | 0.781141 | High | High |
| TLCFTLKRKT | 0.17 | 0.809793 | Low | High |
| LCFTLKRKTE | 0.01 | 0.809793 | Low | High |
| ORF8 | ||||
| SKWYIRVGAR | 0.06 | 0.354214 | Low | High |
| KWYIRVGARK | 0.15 | 0.337595 | Low | High |
| YIRVGARKSA | 0.13 | 0.276083 | Low | High |
* Higher scores show more possibility of cell-penetrating potential.
Uptake efficiency analysis of the identified CPPs
The uptake efficiency of the predicted CPPs was evaluated using two different web servers such as CPPred-RF and MLCPP. These web servers classify CPPs in two categories: high or low uptake efficiency (Table 1).
Calculation of peptide properties
Various physicochemical characteristics of peptides were recognized by diverse web servers such as net charge, pI, MW, amphipathicity, water solubility, hydrophobicity, hydrophobicity ratio, and polar-, non-polar-, uncharged- and charged residues. For instance, a cationic CPP can bind to cell membrane (with negative charge), then can penetrate and deliver cargoes into cells [9]. All of the physicochemical properties of CPPs were determined in Table 2.
Table 2. The properties of peptides determined by diverse web servers and tools.
| Epitope | Net charge | pI | Amphipathicity | MW | Water solubility | Hydrophobicity (H) | Hydrophobic ratio by APD | Polar residues + GLY (n/%) | Nonpolar residues (n/%) | Uncharged residues + GLY | Charged residues |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S protein | |||||||||||
| NLTTRTQLPP | 1.00 | 10.11 | 0.37 | 1140.31 | Poor | 0.379 | 20% | 6/60.00 | 4/40.00 | GLN 1, THR 3, ASN 1, GLY 0 | ARG 1 |
| RFQTLLALHR | 2.50 | 12.01 | 0.76 | 1254.50 | Poor | 0.535 | 50% | 5/50.00 | 5/50.00 | GLN 1, HIS 1, THR 1, GLY 0 | ARG 2 |
| YLQPRTFLLK | 2.00 | 10.01 | 0.74 | 1278.56 | Poor | 0.661 | 40% | 4/40.00 | 6/60.00 | GLN 1, THR 1, GLY 0 | LYS 1, ARG 1 |
| SVYAWNRKRI | 3.00 | 11.01 | 0.86 | 1292.51 | Good | 0.289 | 40% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| YAWNRKRISN | 3.00 | 11.01 | 0.86 | 1307.48 | Good | 0.107 | 30% | 6/60.00 | 4/40.00 | SER 1, ASN 2, GLY 0 | LYS 1, ARG 2 |
| AWNRKRISNC | 3.00 | 10.87 | 0.86 | 1247.45 | Good | 0.165 | 40% | 6/60.00 | 4/40.00 | SER 1, ASN 2, GLY 0 | LYS 1, ARG 2 |
| WNRKRISNCV | 3.00 | 10.87 | 0.86 | 1275.50 | Good | 0.256 | 40% | 6/60.00 | 4/40.00 | SER 1, ASN 2, GLY 0 | LYS 1, ARG 2 |
| RQIAPGQTGK | 2.00 | 11.01 | 0.86 | 1055.20 | Good | 0.065 | 20% | 7/70.00 | 3/30.00 | GLN 2, THR 1, GLY 2 | LYS 1, ARG 1 |
| YNYLYRLFRK | 3.00 | 10.00 | 0.86 | 1435.69 | Good | 0.446 | 30% | 4/40.00 | 6/60.00 | ASN 1, GLY 0 | LYS 1, ARG 2 |
| YLYRLFRKSN | 3.00 | 10.29 | 0.86 | 1435.69 | Good | 0.346 | 30% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| YRLFRKSNLK | 4.00 | 11.10 | 1.22 | 1324.59 | Good | 0.151 | 30% | 6/60.00 | 4/40.00 | SER 1, ASN 1, GLY 0 | LYS 2, ARG 2 |
| RLFRKSNLKP | 4.00 | 12.02 | 1.22 | 1258.53 | Good | 0.127 | 30% | 6/60.00 | 4/40.00 | SER 1, ASN 1, GLY 0 | LYS 2, ARG 2 |
| RKSNLKPFER | 3.00 | 11.01 | 1.35 | 1274.49 | Good | -0.107 | 20% | 7/70.00 | 3/30.00 | SER 1, ASN 1, GLY 0 | LYS 2, ARG 2, GLU 1 |
| KKSTNLVKNK | 4.00 | 10.49 | 1.47 | 1159.40 | Good | -0.202 | 20% | 8/80.00 | 2/20.00 | SER 1, THR 1, ASN 2, GLY 0 | LYS 4 |
| KSTNLVKNKC | 3.00 | 9.81 | 1.10 | 1134.37 | Good | 0.051 | 30% | 7/70.00 | 3/30.00 | SER 1, THR 1, ASN 2, GLY 0 | LYS 3 |
| HADQLTPTWR | 0.50 | 7.10 | 0.52 | 1224.34 | Good | 0.363 | 30% | 6/60.00 | 4/40.00 | GLN 1, HIS 1, THR 2, GLY 0 | ARG 1, ASP 1 |
| YQTQTNSPRR | 2.00 | 10.84 | 0.74 | 1250.34 | Good | -0.090 | 0% | 8/80.00 | 2/20.00 | GLN 2, SER 1, THR 2, ASN 1, GLY 0 | ARG 2 |
| TQTNSPRRAR | 3.00 | 12.31 | 0.86 | 1186.30 | Good | -0.234 | 10% | 8/80.00 | 2/20.00 | GLN 1, SER 1, THR 2, ASN 1, GLY 0 | ARG 3 |
| TNSPRRARSV | 3.00 | 12.31 | 0.74 | 1143.27 | Good | -0.120 | 20% | 7/70.00 | 3/30.00 | SER 2, THR 1, ASN 1, GLY 0 | ARG 3 |
| NSPRRARSVA | 3.00 | 12.31 | 0.74 | 1113.25 | Good | -0.115 | 30% | 6/60.00 | 4/40.00 | SER 2, ASN 1, GLY 0 | ARG 3 |
| PRRARSVASQ | 3.00 | 12.31 | 0.86 | 1127.27 | Good | -0.077 | 30% | 6/60.00 | 4/40.00 | GLN 1, SER 2, GLY 0 | ARG 3 |
| KQIYKTPPIK | 3.00 | 10.01 | 1.23 | 1215.50 | Good | 0.307 | 20% | 5/50.00 | 5/50.00 | GLN 1, THR 1, GLY 0 | LYS 3 |
| SQILPDPSKP | 0.00 | 6.19 | 0.49 | 1081.23 | Good | 0.360 | 20% | 5/50.00 | 5/50.00 | GLN 1, SER 2, GLY 0 | LYS 1, ASP 1 |
| RLITGRLQSL | 2.00 | 12.01 | 0.61 | 1156.39 | Good | 0.488 | 40% | 6/60.00 | 4/40.00 | GLN 1, SER 1, THR 1, GLY 1 | ARG 2 |
| M protein | |||||||||||
| NRNRFLYIIK | 3.00 | 11.01 | 0.86 | 1336.60 | Good | 0.384 | 40% | 5/50.00 | 5/50.00 | ASN 2, GLY 0 | LYS 1, ARG 2 |
| RNRFLYIIKL | 3.00 | 11.01 | 0.86 | 1335.66 | Good | 0.614 | 50% | 4/40.00 | 6/60.00 | ASN 1, GLY 0 | LYS 1, ARG 2 |
| YIIKLIFLWL | 1.00 | 8.94 | 0.37 | 1321.71 | Poor | 1.451 | 80% | 1/10.00 | 9/90.00 | GLY 0 | LYS 1 |
| KLIFLWLLWP | 1.00 | 9.11 | 0.37 | 1328.71 | Poor | 1.462 | 80% | 1/10.00 | 9/90.00 | GLY 0 | LYS 1 |
| FIASFRLFAR | 2.00 | 12.01 | 0.49 | 1227.48 | Poor | 0.743 | 70% | 3/30.00 | 7/70.00 | SER 1, GLY 0 | ARG 2 |
| ASFRLFARTR | 3.00 | 12.31 | 0.74 | 1224.43 | Good | 0.309 | 50% | 5/50.00 | 5/50.00 | SER 1, THR 1, GLY 0 | ARG 3 |
| SFRLFARTRS | 3.00 | 12.31 | 0.74 | 1240.43 | Good | 0.274 | 40% | 6/60.00 | 4/40.00 | SER 2, THR 1, GLY 0 | ARG 3 |
| FRLFARTRSM | 3.00 | 12.31 | 0.74 | 1284.55 | Good | 0.401 | 50% | 5/50.00 | 5/50.00 | SER 1, THR 1, GLY 0 | ARG 3 |
| RLFARTRSMW | 3.00 | 12.31 | 0.74 | 1323.59 | Good | 0.447 | 50% | 5/50.00 | 5/50.00 | SER 1, THR 1, GLY 0 | ARG 3 |
| FARTRSMWSF | 2.00 | 12.01 | 0.49 | 1323.59 | Poor | 0.553 | 50% | 5/50.00 | 5/50.00 | SER 2, THR 1, GLY 0 | ARG 2 |
| HGTILTRPLL | 1.50 | 10.11 | 0.39 | 1120.36 | Poor | 0.726 | 40% | 5/50.00 | 5/50.00 | HIS 1, THR 2, GLY 1 | ARG 1 |
| GAVILRGHLR | 2.50 | 12.01 | 0.64 | 1091.33 | Good | 0.484 | 50% | 5/50.00 | 5/50.00 | HIS 1, GLY 2 | ARG 2 |
| RIAGHHLGRC | 3.00 | 10.38 | 0.78 | 1119.32 | Good | 0.359 | 40% | 6/60.00 | 4/40.00 | HIS 2, GLY 2 | ARG 2 |
| YSRYRIGNYK | 3.00 | 10.00 | 0.86 | 1319.49 | Good | 0.103 | 10% | 6/60.00 | 4/40.00 | SER 1, ASN 1, GLY 1 | LYS 1, ARG 2 |
| N protein | |||||||||||
| PQNQRNAPRI | 2.00 | 12.01 | 0.74 | 1193.33 | Good | -0.011 | 20% | 6/60.00 | 4/40.00 | GLN 2, ASN 2, GLY 0 | ARG 2 |
| ERSGARSKQR | 3.00 | 11.72 | 1.35 | 1174.29 | Good | -0.465 | 10% | 9/90.00 | 1/10.00 | GLN 1, SER 2, GLY 1 | LYS 1, ARG 3, GLU 1 |
| RSGARSKQRR | 5.00 | 12.48 | 1.47 | 1201.36 | Good | -0.502 | 10% | 9/90.00 | 1/10.00 | GLN 1, SER 2, GLY 1 | LYS 1, ARG 4 |
| SGARSKQRRP | 4.00 | 12.31 | 1.23 | 1142.29 | Good | -0.329 | 10% | 8/80.00 | 2/20.00 | GLN 1, SER 2, GLY 1 | LYS 1, ARG 3 |
| GARSKQRRPQ | 4.00 | 12.31 | 1.35 | 1183.34 | Good | -0.347 | 10% | 8/80.00 | 2/20.00 | GLN 2, SER 1, GLY 1 | LYS 1, ARG 3 |
| ARSKQRRPQG | 4.00 | 12.31 | 1.35 | 1183.34 | Good | -0.347 | 10% | 8/80.00 | 2/20.00 | GLN 2, SER 1, GLY 1 | LYS 1, ARG 3 |
| RSKQRRPQGL | 4.00 | 12.31 | 1.35 | 1225.42 | Good | -0.208 | 10% | 8/80.00 | 2/20.00 | GLN 2, SER 1, GLY 1 | LYS 1, ARG 3 |
| SKQRRPQGLP | 3.00 | 12.01 | 1.11 | 1166.35 | Good | -0.035 | 10% | 7/70.00 | 3/30.00 | GLN 2, SER 1, GLY 1 | LYS 1, ARG 2 |
| KQRRPQGLPN | 3.00 | 12.01 | 1.11 | 1193.38 | Good | -0.091 | 10% | 7/70.00 | 3/30.00 | GLN 2, ASN 1, GLY 1 | LYS 1, ARG 2 |
| RRPQGLPNNT | 2.00 | 12.01 | 0.61 | 1152.28 | Good | -0.004 | 10% | 7/70.00 | 3/30.00 | GLN 1, THR 1, ASN 2, GLY 1 | ARG 2 |
| QIGYYRRATR | 3.00 | 10.91 | 0.86 | 1283.46 | Good | 0.104 | 20% | 6/60.00 | 4/40.00 | GLN 1, THR 1, GLY 1 | ARG 3 |
| IGYYRRATRR | 4.00 | 11.56 | 0.98 | 1311.51 | Good | 0.025 | 20% | 6/60.00 | 4/40.00 | THR 1, GLY 1 | ARG 4 |
| GYYRRATRRI | 4.00 | 11.56 | 0.98 | 1311.51 | Good | 0.025 | 20% | 6/60.00 | 4/40.00 | THR 1, GLY 1 | ARG 4 |
| YYRRATRRIR | 5.00 | 11.84 | 1.23 | 1410.65 | Good | -0.076 | 20% | 6/60.00 | 4/40.00 | THR 1, GLY 0 | ARG 5 |
| YRRATRRIRG | 5.00 | 12.18 | 1.23 | 1410.65 | Good | -0.172 | 20% | 7/70.00 | 3/30.00 | THR 1, GLY 1 | ARG 5 |
| RRATRRIRGG | 5.00 | 12.61 | 1.23 | 1198.40 | Good | -0.268 | 20% | 8/80.00 | 2/20.00 | THR 1, GLY 2 | ARG 5 |
| RATRRIRGGD | 3.00 | 12.01 | 0.98 | 1157.30 | Good | -0.244 | 20% | 8/80.00 | 2/20.00 | THR 1, GLY 2 | ARG 4, ASP 1 |
| TRRIRGGDGK | 3.00 | 11.72 | 1.10 | 1115.26 | Good | -0.273 | 30% | 9/90.00 | 1/10.00 | THR 1, GLY 3 | LYS 1, ARG 3, ASP 1 |
| RIRGGDGKMK | 3.00 | 11.01 | 1.22 | 1117.34 | Good | -0.174 | 20% | 8/80.00 | 2/20.00 | GLY 3 | LYS 2, ARG 2, ASP 1 |
| GKMKDLSPRW | 2.00 | 10.01 | 0.98 | 1217.46 | Good | 0.210 | 30% | 6/60.00 | 4/40.00 | SER 1, GLY 1 | LYS 2, ARG 1, ASP 1 |
| SQASSRSSSR | 2.00 | 12.01 | 0.61 | 1052.07 | Good | -0.217 | 10% | 9/90.00 | 1/10.00 | GLN 1, SER 6, GLY 0 | ARG 2 |
| ASSRSSSRSR | 3.00 | 12.31 | 0.74 | 1080.13 | Good | -0.296 | 10% | 9/90.00 | 1/10.00 | SER 6, GLY 0 | ARG 3 |
| SSRSSSRSRN | 3.00 | 12.31 | 0.74 | 1123.15 | Good | -0.387 | 0% | 10/100.00 | 0/0.00 | SER 6, ASN 1, GLY 0 | ARG 3 |
| SRSSSRSRNS | 3.00 | 12.31 | 0.74 | 1123.15 | Good | -0.387 | 0% | 10/100.00 | 0/0.00 | SER 6, ASN 1, GLY 0 | ARG 3 |
| RSSSRSRNSS | 3.00 | 12.31 | 0.74 | 1123.15 | Good | -0.387 | 0% | 10/100.00 | 0/0.00 | SER 6, ASN 1, GLY 0 | ARG 3 |
| SSSRSRNSSR | 3.00 | 12.31 | 0.74 | 1123.15 | Good | -0.387 | 0% | 10/100.00 | 0/0.00 | SER 6, ASN 1, GLY 0 | ARG 3 |
| SSRSRNSSRN | 3.00 | 12.31 | 0.74 | 1150.18 | Good | -0.443 | 0% | 10/100.00 | 0/0.00 | SER 5, ASN 2, GLY 0 | ARG 3 |
| RSRNSSRNST | 3.00 | 12.31 | 0.74 | 1164.20 | Good | -0.413 | 0% | 10/100.00 | 0/0.00 | SER 4, THR 1, ASN 2, GLY 0 | ARG 3 |
| GSSRGTSPAR | 2.00 | 12.01 | 0.49 | 975.03 | Good | -0.085 | 10% | 8/80.00 | 2/20.00 | SER 3, THR 1, GLY 2 | ARG 2 |
| AALALLLLDR | 0.00 | 6.19 | 0.25 | 1068.33 | Poor | 0.765 | 80% | 2/20.00 | 8/80.00 | GLY 0 | ARG 1, ASP 1 |
| ALALLLLDRL | 0.00 | 6.19 | 0.25 | 1110.41 | Poor | 0.904 | 80% | 2/20.00 | 8/80.00 | GLY 0 | ARG 1, ASP 1 |
| KKSAAEASKK | 3.00 | 10.01 | 1.59 | 1047.22 | Good | -0.375 | 30% | 7/70.00 | 3/30.00 | SER 2, GLY 0 | LYS 4, GLU 1 |
| KSAAEASKKP | 2.00 | 9.72 | 1.23 | 1016.16 | Good | -0.204 | 30% | 6/60.00 | 4/40.00 | SER 2, GLY 0 | LYS 3, GLU 1 |
| SAAEASKKPR | 2.00 | 10.01 | 1.11 | 1044.18 | Good | -0.206 | 30% | 6/60.00 | 4/40.00 | SER 2, GLY 0 | LYS 2, ARG 1, GLU 1 |
| AAEASKKPRQ | 2.00 | 10.01 | 1.23 | 1085.23 | Good | -0.224 | 30% | 6/60.00 | 4/40.00 | GLN 1, SER 1, GLY 0 | LYS 2, ARG 1, GLU 1 |
| AEASKKPRQK | 3.00 | 10.30 | 1.60 | 1142.32 | Good | -0.354 | 20% | 7/70.00 | 3/30.00 | GLN 1, SER 1, GLY 0 | LYS 3, ARG 1, GLU 1 |
| EASKKPRQKR | 4.00 | 11.10 | 1.84 | 1227.43 | Good | -0.486 | 10% | 8/80.00 | 2/20.00 | GLN 1, SER 1, GLY 0 | LYS 3, ARG 2, GLU 1 |
| ASKKPRQKRT | 5.00 | 12.03 | 1.72 | 1199.42 | Good | -0.396 | 10% | 8/80.00 | 2/20.00 | GLN 1, SER 1, THR 1, GLY 0 | LYS 3, ARG 2 |
| SKKPRQKRTA | 5.00 | 12.03 | 1.72 | 1199.42 | Good | -0.396 | 10% | 8/80.00 | 2/20.00 | GLN 1, SER 1, THR 1, GLY 0 | LYS 3, ARG 2 |
| KKPRQKRTAT | 5.00 | 12.03 | 1.72 | 1213.45 | Good | -0.366 | 10% | 8/80.00 | 2/20.00 | GLN 1, THR 2, GLY 0 | LYS 3, ARG 2 |
| KPRQKRTATK | 5.00 | 12.03 | 1.72 | 1213.45 | Good | -0.366 | 10% | 8/80.00 | 2/20.00 | GLN 1, THR 2, GLY 0 | LYS 3, ARG 2 |
| PRQKRTATKA | 4.00 | 12.02 | 1.35 | 1156.35 | Good | -0.236 | 20% | 7/70.00 | 3/30.00 | GLN 1, THR 2, GLY 0 | LYS 2, ARG 2 |
| RQKRTATKAY | 4.00 | 11.10 | 1.35 | 1222.41 | Good | -0.212 | 20% | 7/70.00 | 3/30.00 | GLN 1, THR 2, GLY 0 | LYS 2, ARG 2 |
| RQGTDYKHWP | 1.50 | 8.94 | 0.88 | 1287.40 | Good | 0.133 | 10% | 7/70.00 | 3/30.00 | GLN 1, HIS 1, THR 1, GLY 1 | LYS 1, ARG 1, ASP 1 |
| FPPTEPKKDK | 1.00 | 8.83 | 1.23 | 1186.37 | Good | -0.017 | 10% | 6/60.00 | 4/40.00 | THR 1, GLY 0 | LYS 3, GLU 1, ASP 1 |
| PPTEPKKDKK | 2.00 | 9.55 | 1.59 | 1167.37 | Good | -0.295 | 0% | 7/70.00 | 3/30.00 | THR 1, GLY 0 | LYS 4, GLU 1, ASP 1 |
| PTEPKKDKKK | 3.00 | 9.84 | 1.96 | 1198.43 | Good | -0.466 | 0% | 8/80.00 | 2/20.00 | THR 1, GLY 0 | LYS 5, GLU 1, ASP 1 |
| TEPKKDKKKK | 4.00 | 10.01 | 2.33 | 1229.49 | Good | -0.637 | 0% | 9/90.00 | 1/10.00 | THR 1, GLY 0 | LYS 6, GLU 1, ASP 1 |
| EPKKDKKKKA | 4.00 | 10.01 | 2.33 | 1199.46 | Good | -0.632 | 10% | 8/80.00 | 2/20.00 | GLY 0 | LYS 6, GLU 1, ASP 1 |
| PKKDKKKKAD | 4.00 | 10.01 | 2.20 | 1185.43 | Good | -0.645 | 10% | 8/80.00 | 2/20.00 | GLY 0 | LYS 6, ASP 2 |
| KKDKKKKADE | 3.00 | 9.71 | 2.33 | 1217.43 | Good | -0.781 | 10% | 9/90.00 | 1/10.00 | GLY 0 | LYS 6, GLU 1, ASP 2 |
| TQALPQRQKK | 3.00 | 11.17 | 1.35 | 1197.40 | Good | -0.066 | 20% | 7/70.00 | 3/30.00 | GLN 3, THR 1, GLY 0 | LYS 2, ARG 1 |
| ALPQRQKKQQ | 3.00 | 11.17 | 1.48 | 1224.43 | Good | -0.114 | 20% | 7/70.00 | 3/30.00 | GLN 4, GLY 0 | LYS 2, ARG 1 |
| ORF3a | |||||||||||
| SASKIITLKK | 3.00 | 10.31 | 1.10 | 1088.36 | Good | 0.282 | 40% | 6/60.00 | 4/40.00 | SER 2, THR 1, GLY 0 | LYS 3 |
| ASKIITLKKR | 4.00 | 11.27 | 1.35 | 1157.47 | Good | 0.185 | 40% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 0 | LYS 3, ARG 1 |
| SKIITLKKRW | 4.00 | 11.27 | 1.35 | 1272.60 | Good | 0.379 | 40% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 0 | LYS 3, ARG 1 |
| KIITLKKRWQ | 4.00 | 11.27 | 1.47 | 1313.65 | Good | 0.361 | 40% | 6/60.00 | 4/40.00 | GLN 1, THR 1, GLY 0 | LYS 3, ARG 1 |
| IITLKKRWQL | 3.00 | 11.17 | 1.10 | 1298.64 | Good | 0.630 | 50% | 5/50.00 | 5/50.00 | GLN 1, THR 1, GLY 0 | LYS 2, ARG 1 |
| KKRWQLALSK | 4.00 | 11.27 | 1.47 | 1257.55 | Good | 0.172 | 40% | 6/60.00 | 4/40.00 | GLN 1, SER 1, GLY 0 | LYS 3, ARG 1 |
| KRWQLALSKG | 3.00 | 11.17 | 1.10 | 1186.42 | Good | 0.271 | 40% | 6/60.00 | 4/40.00 | GLN 1, SER 1, GLY 1 | LYS 2, ARG 1 |
| VRIIMRLWLC | 2.00 | 10.38 | 0.49 | 1302.72 | Poor | 1.122 | 80% | 2/20.00 | 8/80.00 | GLY 0 | ARG 2 |
| RIIMRLWLCW | 2.00 | 10.38 | 0.49 | 1389.80 | Poor | 1.225 | 80% | 2/20.00 | 8/80.00 | GLY 0 | ARG 2 |
| IIMRLWLCWK | 2.00 | 9.55 | 0.61 | 1361.79 | Poor | 1.227 | 80% | 2/20.00 | 8/80.00 | GLY 0 | LYS 1, ARG 1 |
| IMRLWLCWKC | 2.00 | 9.03 | 0.61 | 1351.77 | Poor | 1.201 | 80% | 2/20.00 | 8/80.00 | GLY 0 | LYS 1, ARG 1 |
| MRLWLCWKCR | 3.00 | 9.72 | 0.86 | 1394.80 | Good | 0.920 | 70% | 3/30.00 | 7/70.00 | GLY 0 | LYS 1, ARG 2 |
| RLWLCWKCRS | 3.00 | 9.72 | 0.86 | 1350.68 | Good | 0.793 | 60% | 4/40.00 | 6/60.00 | SER 1, GLY 0 | LYS 1, ARG 2 |
| LWLCWKCRSK | 3.00 | 9.53 | 0.98 | 1322.66 | Good | 0.795 | 60% | 4/40.00 | 6/60.00 | SER 1, GLY 0 | LYS 2, ARG 1 |
| WLCWKCRSKN | 3.00 | 9.53 | 0.98 | 1323.61 | Good | 0.565 | 50% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 2, ARG 1 |
| LCWKCRSKNP | 3.00 | 9.53 | 0.98 | 1234.51 | Good | 0.412 | 40% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 2, ARG 1 |
| CWKCRSKNPL | 3.00 | 9.53 | 0.98 | 1234.51 | Good | 0.412 | 40% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 2, ARG 1 |
| KCRSKNPLLY | 3.00 | 9.80 | 0.98 | 1221.49 | Good | 0.299 | 30% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 2, ARG 1 |
| Orf1ab | |||||||||||
| IKRSDARTAP | 2.00 | 10.84 | 0.86 | 1114.27 | Good | -0.042 | 30% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 0 | LYS 1, ARG 2, ASP 1 |
| KRSDARTAPH | 2.50 | 10.84 | 1.00 | 1138.25 | Good | -0.209 | 20% | 7/70.00 | 3/30.00 | HIS 1, SER 1, THR 1, GLY 0 | LYS 1, ARG 2, ASP 1 |
| PVAYRKVLLR | 3.00 | 11.01 | 0.86 | 1214.52 | Good | 0.482 | 50% | 3/30.00 | 7/70.00 | GLY 0 | LYS 1, ARG 2 |
| VAYRKVLLRK | 4.00 | 11.10 | 1.22 | 1245.58 | Good | 0.311 | 50% | 4/40.00 | 6/60.00 | GLY 0 | LYS 2, ARG 2 |
| AYRKVLLRKN | 4.00 | 11.10 | 1.22 | 1260.55 | Good | 0.129 | 40% | 5/50.00 | 5/50.00 | ASN 1, GLY 0 | LYS 2, ARG 2 |
| YRKVLLRKNG | 4.00 | 11.10 | 1.22 | 1246.52 | Good | 0.098 | 30% | 6/60.00 | 4/40.00 | ASN 1, GLY 1 | LYS 2, ARG 2 |
| RKVLLRKNGN | 4.00 | 12.02 | 1.22 | 1197.45 | Good | -0.058 | 30% | 7/70.00 | 3/30.00 | ASN 2, GLY 1 | LYS 2, ARG 2 |
| KVLLRKNGNK | 4.00 | 11.27 | 1.35 | 1169.44 | Good | -0.056 | 30% | 7/70.00 | 3/30.00 | ASN 2, GLY 1 | LYS 3, ARG 1 |
| FEIKLAKKFD | 1.00 | 8.83 | 1.23 | 1238.49 | Good | 0.301 | 50% | 5/50.00 | 5/50.00 | GLY 0 | LYS 3, GLU 1, ASP 1 |
| KTIQPRVEKK | 3.00 | 10.30 | 1.60 | 1226.49 | Good | -0.084 | 20% | 7/70.00 | 3/30.00 | GLN 1, THR 1, GLY 0 | LYS 3, ARG 1, GLU 1 |
| TIQPRVEKKK | 3.00 | 10.30 | 1.60 | 1226.49 | Good | -0.084 | 20% | 7/70.00 | 3/30.00 | GLN 1, THR 1, GLY 0 | LYS 3, ARG 1, GLU 1 |
| IQPRVEKKKL | 3.00 | 10.30 | 1.60 | 1238.54 | Good | 0.060 | 30% | 6/60.00 | 4/40.00 | GLN 1, GLY 0 | LYS 3, ARG 1, GLU 1 |
| SGLKTILRKG | 3.00 | 11.17 | 0.98 | 1072.32 | Good | 0.243 | 30% | 7/70.00 | 3/30.00 | SER 1, THR 1, GLY 2 | LYS 2, ARG 1 |
| LKTILRKGGR | 4.00 | 12.02 | 1.22 | 1141.43 | Good | 0.146 | 30% | 7/70.00 | 3/30.00 | THR 1, GLY 2 | LYS 2, ARG 2 |
| KTILRKGGRT | 4.00 | 12.02 | 1.22 | 1129.37 | Good | 0.002 | 20% | 8/80.00 | 2/20.00 | THR 2, GLY 2 | LYS 2, ARG 2 |
| GNFKVTKGKA | 3.00 | 10.31 | 1.10 | 1049.24 | Good | 0.001 | 30% | 7/70.00 | 3/30.00 | THR 1, ASN 1, GLY 2 | LYS 3 |
| FKVTKGKAKK | 5.00 | 10.61 | 1.84 | 1134.43 | Good | -0.137 | 30% | 7/70.00 | 3/30.00 | THR 1, GLY 1 | LYS 5 |
| KVTKGKAKKG | 5.00 | 10.61 | 1.84 | 1044.31 | Good | -0.316 | 20% | 8/80.00 | 2/20.00 | THR 1, GLY 2 | LYS 5 |
| KGKAKKGAWN | 4.00 | 10.49 | 1.47 | 1087.29 | Good | -0.169 | 30% | 7/70.00 | 3/30.00 | ASN 1, GLY 2 | LYS 4 |
| GGAKLKALNL | 2.00 | 10.02 | 0.73 | 984.21 | Good | 0.314 | 50% | 5/50.00 | 5/50.00 | ASN 1, GLY 2 | LYS 2 |
| SKGLYRKCVK | 4.00 | 10.04 | 1.35 | 1181.47 | Good | 0.140 | 30% | 6/60.00 | 4/40.00 | SER 1, GLY 1 | LYS 3, ARG 1 |
| KGLYRKCVKS | 4.00 | 10.04 | 1.35 | 1181.47 | Good | 0.140 | 30% | 6/60.00 | 4/40.00 | SER 1, GLY 1 | LYS 3, ARG 1 |
| GLYRKCVKSR | 4.00 | 10.32 | 1.22 | 1209.48 | Good | 0.138 | 30% | 6/60.00 | 4/40.00 | SER 1, GLY 1 | LYS 2, ARG 2 |
| GLLMPLKAPK | 2.00 | 10.02 | 0.73 | 1067.40 | Good | 0.610 | 50% | 3/30.00 | 7/70.00 | GLY 1 | LYS 2 |
| QRKQDDKKIK | 3.00 | 10.01 | 1.96 | 1286.50 | Good | -0.515 | 10% | 9/90.00 | 1/10.00 | GLN 2, GLY 0 | LYS 4, ARG 1, ASP 2 |
| RKQDDKKIKA | 3.00 | 10.01 | 1.84 | 1229.45 | Good | -0.462 | 20% | 8/80.00 | 2/20.00 | GLN 1, GLY 0 | LYS 4, ARG 1, ASP 2 |
| KQDDKKIKAC | 2.00 | 9.17 | 1.59 | 1176.40 | Good | -0.207 | 30% | 7/70.00 | 3/30.00 | GLN 1, GLY 0 | LYS 4, ASP 2 |
| DITFLKKDAP | 0.00 | 6.31 | 0.73 | 1147.34 | Good | 0.306 | 40% | 5/50.00 | 5/50.00 | THR 1, GLY 0 | LYS 2, ASP 2 |
| MLAKALRKVP | 3.00 | 11.17 | 0.98 | 1126.48 | Good | 0.420 | 60% | 3/30.00 | 7/70.00 | GLY 0 | LYS 2, ARG 1 |
| LAKALRKVPT | 3.00 | 11.17 | 0.98 | 1096.38 | Good | 0.323 | 50% | 4/40.00 | 6/60.00 | THR 1, GLY 0 | LYS 2, ARG 1 |
| EAKTVLKKCK | 3.00 | 9.65 | 1.59 | 1147.45 | Good | 0.043 | 40% | 6/60.00 | 4/40.00 | THR 1, GLY 0 | LYS 4, GLU 1 |
| AKTVLKKCKS | 4.00 | 10.05 | 1.47 | 1105.41 | Good | 0.103 | 40% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 0 | LYS 4 |
| KTVLKKCKSA | 4.00 | 10.05 | 1.47 | 1105.41 | Good | 0.103 | 40% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 0 | LYS 4 |
| KSAFYILPSI | 1.00 | 8.94 | 0.37 | 1138.37 | Poor | 0.801 | 50% | 3/30.00 | 7/70.00 | SER 2, GLY 0 | LYS 1 |
| KAIVSTIQRK | 3.00 | 11.17 | 1.10 | 1143.40 | Good | 0.214 | 40% | 6/60.00 | 4/40.00 | GLN 1, SER 1, THR 1, GLY 0 | LYS 2, ARG 1 |
| STIQRKYKGI | 3.00 | 10.30 | 1.10 | 1193.41 | Good | 0.157 | 20% | 7/70.00 | 3/30.00 | GLN 1, SER 1, THR 1, GLY 1 | LYS 2, ARG 1 |
| TIQRKYKGIK | 4.00 | 10.47 | 1.47 | 1234.5 | Good | 0.062 | 20% | 7/70.00 | 3/30.00 | GLN 1, THR 1, GLY 1 | LYS 3, ARG 1 |
| IQRKYKGIKI | 4.00 | 10.47 | 1.47 | 1246.56 | Good | 0.216 | 30% | 6/60.00 | 4/40.00 | GLN 1, GLY 1 | LYS 3, ARG 1 |
| GARFYFYTSK | 2.00 | 9.72 | 0.61 | 1239.40 | Poor | 0.403 | 30% | 5/50.00 | 5/50.00 | SER 1, THR 1, GLY 1 | LYS 1, ARG 1 |
| ARYMRSLKVP | 3.00 | 11.01 | 0.86 | 1220.51 | Good | 0.309 | 40% | 4/40.00 | 6/60.00 | SER 1, GLY 0 | LYS 1, ARG 2 |
| GIEFLKRGDK | 1.00 | 8.93 | 1.11 | 1162.35 | Good | 0.089 | 30% | 7/70.00 | 3/30.00 | GLY 2 | LYS 2, ARG 1, GLU 1, ASP 1 |
| DNLKTLLSLR | 1.00 | 9.10 | 0.61 | 1172.39 | Good | 0.365 | 40% | 6/60.00 | 4/40.00 | SER 1, THR 1, ASN 1, GLY 0 | LYS 1, ARG 1, ASP 1 |
| YMSALNHTKK | 2.50 | 9.72 | 0.88 | 1192.41 | Good | 0.197 | 30% | 6/60.00 | 4/40.00 | HIS 1, SER 1, THR 1, ASN 1, GLY 0 | LYS 2 |
| SALNHTKKWK | 3.50 | 10.31 | 1.25 | 1212.42 | Good | 0.104 | 30% | 7/70.00 | 3/30.00 | HIS 1, SER 1, THR 1, ASN 1, GLY 0 | LYS 3 |
| ALNHTKKWKY | 3.50 | 10.01 | 1.25 | 1288.52 | Good | 0.204 | 30% | 6/60.00 | 4/40.00 | HIS 1, THR 1, ASN 1, GLY 0 | LYS 3 |
| LNHTKKWKYP | 3.50 | 10.01 | 1.25 | 1314.55 | Good | 0.245 | 20% | 6/60.00 | 4/40.00 | HIS 1, THR 1, ASN 1, GLY 0 | LYS 3 |
| NHTKKWKYPQ | 3.50 | 10.01 | 1.37 | 1329.52 | Good | 0.053 | 10% | 7/70.00 | 3/30.00 | GLN 1, HIS 1, THR 1, ASN 1, GLY 0 | LYS 3 |
| HTKKWKYPQV | 3.50 | 10.01 | 1.37 | 1314.55 | Good | 0.235 | 20% | 6/60.00 | 4/40.00 | GLN 1, HIS 1, THR 1, GLY 0 | LYS 3 |
| KKPASRELKV | 3.00 | 10.30 | 1.47 | 1155.41 | Good | -0.071 | 30% | 6/60.00 | 4/40.00 | SER 1, GLY 0 | LYS 3, ARG 1, GLU 1 |
| KPASRELKVT | 2.00 | 10.01 | 1.11 | 1128.34 | Good | 0.054 | 30% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 0 | LYS 2, ARG 1, GLU 1 |
| YTPSFKKGAK | 3.00 | 10.01 | 1.10 | 1126.32 | Good | 0.103 | 20% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 1 | LYS 3 |
| PSFKKGAKLL | 3.00 | 10.31 | 1.10 | 1088.36 | Good | 0.321 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | LYS 3 |
| FKKGAKLLHK | 4.50 | 10.49 | 1.61 | 1169.48 | Good | 0.167 | 40% | 6/60.00 | 4/40.00 | HIS 1, GLY 1 | LYS 4 |
| KKGAKLLHKP | 4.50 | 10.49 | 1.61 | 1119.42 | Good | 0.060 | 30% | 6/60.00 | 4/40.00 | HIS 1, GLY 1 | LYS 4 |
| KGAKLLHKPI | 3.50 | 10.31 | 1.25 | 1104.41 | Good | 0.339 | 40% | 5/50.00 | 5/50.00 | HIS 1, GLY 1 | LYS 3 |
| WCIRCLWSTK | 2.00 | 9.03 | 0.61 | 1295.60 | Poor | 0.930 | 60% | 4/40.00 | 6/60.00 | SER 1, THR 1, GLY 0 | LYS 1, ARG 1 |
| CIRCLWSTKP | 2.00 | 9.03 | 0.61 | 1206.50 | Poor | 0.777 | 50% | 4/40.00 | 6/60.00 | SER 1, THR 1, GLY 0 | LYS 1, ARG 1 |
| ANYAKPFLNK | 2.00 | 9.7 | 0.73 | 1165.36 | Good | 0.261 | 33% | 4/40.00 | 6/60.00 | ASN 2, GLY 0 | LYS 2 |
| TNIVTRCLNR | 2.00 | 10.38 | 0.49 | 1189.40 | Good | 0.356 | 40% | 6/60.00 | 4/40.00 | THR 2, ASN 2, GLY 0 | ARG 2 |
| CTFTRSTNSR | 2.00 | 10.38 | 0.49 | 1172.29 | Good | 0.141 | 20% | 8/80.00 | 2/20.00 | SER 2, THR 3, ASN 1, GLY 0 | ARG 2 |
| TCMMCYKRNR | 3.00 | 9.53 | 0.86 | 1305.64 | Good | 0.315 | 40% | 5/50.00 | 5/50.00 | THR 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| MCYKRNRATR | 4.00 | 10.92 | 1.10 | 1298.56 | Good | -0.032 | 30% | 6/60.00 | 4/40.00 | THR 1, ASN 1, GLY 0 | LYS 1, ARG 3 |
| CYKRNRATRV | 4.00 | 10.92 | 1.10 | 1266.49 | Good | -0.033 | 30% | 6/60.00 | 4/40.00 | THR 1, ASN 1, GLY 0 | LYS 1, ARG 3 |
| YKRNRATRVE | 3.00 | 10.91 | 1.23 | 1292.46 | Good | -0.251 | 20% | 7/70.00 | 3/30.00 | THR 1, ASN 1, GLY 0 | LYS 1, ARG 3, GLU 1 |
| KRNRATRVEC | 3.00 | 10.77 | 1.23 | 1232.43 | Good | -0.193 | 30% | 7/70.00 | 3/30.00 | THR 1, ASN 1, GLY 0 | LYS 1, ARG 3, GLU 1 |
| RNRATRVECT | 2.00 | 10.29 | 0.86 | 1205.36 | Good | -0.068 | 30% | 7/70.00 | 3/30.00 | THR 2, ASN 1, GLY 0 | ARG 3, GLU 1 |
| RDLSLQFKRP | 2.00 | 10.84 | 0.98 | 1259.47 | Good | 0.187 | 30% | 6/60.00 | 4/40.00 | GLN 1, SER 1, GLY 0 | LYS 1, ARG 2, ASP 1 |
| SLQFKRPINP | 2.00 | 11.01 | 0.74 | 1199.42 | Good | 0.387 | 30% | 5/50.00 | 5/50.00 | GLN 1, SER 1, ASN 1, GLY 0 | LYS 1, ARG 1 |
| HNIALIWNVK | 1.50 | 9.11 | 0.51 | 1207.44 | Poor | 0.702 | 60% | 4/40.00 | 6/60.00 | HIS 1, ASN 2, GLY 0 | LYS 1 |
| LSEQLRKQIR | 2.00 | 10.84 | 1.23 | 1270.50 | Good | 0.107 | 30% | 7/70.00 | 3/30.00 | GLN 2, SER 1, GLY 0 | LYS 1, ARG 2, GLU 1 |
| QLRKQIRSAA | 3.00 | 12.01 | 1.11 | 1170.38 | Good | 0.063 | 40% | 6/60.00 | 4/40.00 | GLN 2, SER 1, GLY 0 | LYS 1, ARG 2 |
| LRKQIRSAAK | 4.00 | 12.02 | 1.35 | 1170.42 | Good | -0.014 | 40% | 6/60.00 | 4/40.00 | GLN 1, SER 1, GLY 0 | LYS 2, ARG 2 |
| RKQIRSAAKK | 5.00 | 12.03 | 1.72 | 1185.44 | Good | -0.283 | 30% | 7/70.00 | 3/30.00 | GLN 1, SER 1, GLY 0 | LYS 3, ARG 2 |
| KQIRSAAKKN | 4.00 | 11.27 | 1.47 | 1143.36 | Good | -0.242 | 30% | 7/70.00 | 3/30.00 | GLN 1, SER 1, ASN 1, GLY 0 | LYS 3, ARG 1 |
| QIRSAAKKNN | 3.00 | 11.17 | 1.10 | 1129.29 | Good | -0.203 | 30% | 7/70.00 | 3/30.00 | GLN 1, SER 1, ASN 2, GLY 0 | LYS 2, ARG 1 |
| AAKKNNLPFK | 3.00 | 10.31 | 1.10 | 1130.36 | Good | 0.066 | 40% | 5/50.00 | 5/50.00 | ASN 2, GLY 0 | LYS 3 |
| KKNNLPFKLT | 3.00 | 10.31 | 1.10 | 1202.46 | Good | 0.200 | 30% | 6/60.00 | 4/40.00 | THR 1, ASN 2, GLY 0 | LYS 3 |
| NNWLKQLIKV | 2.00 | 10.02 | 0.86 | 1255.53 | Poor | 0.527 | 50% | 5/50.00 | 5/50.00 | GLN 1, ASN 2, GLY 0 | LYS 2 |
| LAYYFMRFRR | 3.00 | 10.91 | 0.74 | 1422.72 | Good | 0.571 | 50% | 3/30.00 | 7/70.00 | GLY 0 | ARG 3 |
| AYYFMRFRRA | 3.00 | 10.91 | 0.74 | 1380.64 | Good | 0.432 | 50% | 3/30.00 | 7/70.00 | GLY 0 | ARG 3 |
| YYFMRFRRAF | 3.00 | 10.91 | 0.74 | 1456.74 | Poor | 0.580 | 50% | 3/30.00 | 7/70.00 | GLY 0 | ARG 3 |
| FMRFRRAFGE | 2.00 | 11.70 | 0.86 | 1316.55 | Good | 0.324 | 50% | 5/50.00 | 5/50.00 | GLY 1 | ARG 3, GLU 1 |
| MRFRRAFGEY | 2.00 | 10.75 | 0.86 | 1332.55 | Good | 0.241 | 40% | 5/50.00 | 5/50.00 | GLY 1 | ARG 3, GLU 1 |
| KEMYLKLRSD | 1.00 | 8.83 | 1.11 | 1282.53 | Good | 0.115 | 30% | 6/60.00 | 4/40.00 | SER 1, GLY 0 | LYS 2, ARG 1, GLU 1, ASP 1 |
| YNRYLALYNK | 2.00 | 9.55 | 0.61 | 1317.51 | Poor | 0.339 | 30% | 4/40.00 | 6/60.00 | ASN 2, GLY 0 | LYS 1, ARG 1 |
| RYLALYNKYK | 3.00 | 9.83 | 0.98 | 1331.58 | Good | 0.300 | 30% | 4/40.00 | 6/60.00 | ASN 1, GLY 0 | LYS 2, ARG 1 |
| FRKMAFPSGK | 3.00 | 11.17 | 0.98 | 1168.43 | Good | 0.281 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | LYS 2, ARG 1 |
| TANPKTPKYK | 3.00 | 10.01 | 1.10 | 1147.34 | Good | -0.034 | 10% | 6/60.00 | 4/40.00 | THR 2, ASN 1, GLY 0 | LYS 3 |
| ANPKTPKYKF | 3.00 | 10.01 | 1.10 | 1193.41 | Good | 0.119 | 20% | 5/50.00 | 5/50.00 | THR 1, ASN 1, GLY 0 | LYS 3 |
| PKTPKYKFVR | 4.00 | 10.47 | 1.35 | 1263.55 | Good | 0.169 | 20% | 5/50.00 | 5/50.00 | THR 1, GLY 0 | LYS 3, ARG 1 |
| KTPKYKFVRI | 4.00 | 10.47 | 1.35 | 1279.59 | Good | 0.277 | 30% | 5/50.00 | 5/50.00 | THR 1, GLY 0 | LYS 3, ARG 1 |
| RWFLNRFTTT | 2.00 | 12.01 | 0.49 | 1341.54 | Poor | 0.569 | 40% | 6/60.00 | 4/40.00 | THR 3, ASN 1, GLY 0 | ARG 2 |
| FQSAVKRTIK | 3.00 | 11.17 | 1.10 | 1177.41 | Good | 0.213 | 40% | 6/60.00 | 4/40.00 | GLN 1, SER 1, THR 1, GLY 0 | LYS 2, ARG 1 |
| SEVVLKKLKK | 3.00 | 10.01 | 1.59 | 1171.49 | Good | 0.120 | 40% | 6/60.00 | 4/40.00 | SER 1, GLY 0 | LYS 4, GLU 1 |
| VVLKKLKKSL | 4.00 | 10.49 | 1.47 | 1155.53 | Good | 0.354 | 50% | 5/50.00 | 5/50.00 | SER 1, GLY 0 | LYS 4 |
| VLKKLKKSLN | 4.00 | 10.49 | 1.47 | 1170.51 | Good | 0.172 | 40% | 6/60.00 | 4/40.00 | SER 1, ASN 1, GLY 0 | LYS 4 |
| KKLKKSLNVA | 4.00 | 10.49 | 1.47 | 1128.42 | Good | 0.033 | 40% | 6/60.00 | 4/40.00 | SER 1, ASN 1, GLY 0 | LYS 4 |
| DAAMQRKLEK | 1.00 | 8.93 | 1.23 | 1189.40 | Good | -0.107 | 40% | 6/60.00 | 4/40.00 | GLN 1, GLY 0 | LYS 2, ARG 1, GLU 1, ASP 1 |
| AAMQRKLEKM | 2.00 | 10.01 | 1.23 | 1205.51 | Good | 0.093 | 50% | 5/50.00 | 5/50.00 | GLN 1, GLY 0 | LYS 2, ARG 1, GLU 1 |
| MQRKLEKMAD | 1.00 | 8.93 | 1.23 | 1249.52 | Good | -0.015 | 40% | 6/60.00 | 4/40.00 | GLN 1, GLY 0 | LYS 2, ARG 1, GLU 1, ASP 1 |
| YKQARSEDKR | 2.00 | 9.72 | 1.48 | 1280.41 | Good | -0.440 | 10% | 8/80.00 | 2/20.00 | GLN 1, SER 1, GLY 0 | LYS 2, ARG 2, GLU 1, ASP 1 |
| KQARSEDKRA | 2.00 | 10.00 | 1.48 | 1188.31 | Good | -0.505 | 20% | 8/80.00 | 2/20.00 | GLN 1, SER 1, GLY 0 | LYS 2, ARG 2, GLU 1, ASP 1 |
| QARSEDKRAK | 2.00 | 10.00 | 1.48 | 1188.31 | Good | -0.505 | 20% | 8/80.00 | 2/20.00 | GLN 1, SER 1, GLY 0 | LYS 2, ARG 2, GLU 1, ASP 1 |
| MLFTMLRKLD | 1.00 | 9.10 | 0.61 | 1267.62 | Good | 0.684 | 60% | 4/40.00 | 6/60.00 | THR 1, GLY 0 | LYS 1, ARG 1, ASP 1 |
| QDLKWARFPK | 2.00 | 10.01 | 1.10 | 1288.52 | Good | 0.279 | 40% | 5/50.00 | 5/50.00 | GLN 1, GLY 0 | LYS 2, ARG 1, ASP 1 |
| DLKWARFPKS | 2.00 | 10.01 | 0.98 | 1247.46 | Good | 0.297 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 0 | LYS 2, ARG 1, ASP 1 |
| LKWARFPKSD | 2.00 | 10.01 | 0.98 | 1247.46 | Good | 0.297 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 0 | LYS 2, ARG 1, ASP 1 |
| KGFCDLKGKY | 2.00 | 9.17 | 1.10 | 1158.39 | Good | 0.225 | 30% | 6/60.00 | 4/40.00 | GLY 2 | LYS 3, ASP 1 |
| GVSAARLTPC | 1.00 | 8.60 | 0.25 | 974.15 | Poor | 0.501 | 50% | 4/40.00 | 6/60.00 | SER 1, THR 1, GLY 1 | ARG 1 |
| GFAKFLKTNC | 2.00 | 9.36 | 0.73 | 1128.36 | Poor | 0.481 | 50% | 5/50.00 | 5/50.00 | THR 1, ASN 1, GLY 1 | LYS 2 |
| KTNCCRFQEK | 2.00 | 8.98 | 1.23 | 1256.47 | Good | 0.068 | 30% | 7/70.00 | 3/30.00 | GLN 1, THR 1, ASN 1, GLY 0 | LYS 2, ARG 1, GLU 1 |
| PHISRQRLTK | 3.50 | 12.01 | 1.13 | 1235.46 | Good | 0.134 | 20% | 7/70.00 | 3/30.00 | GLN 1, HIS 1, SER 1, THR 1, GLY 0 | LYS 1, ARG 2 |
| HISRQRLTKY | 3.50 | 11.01 | 1.13 | 1301.52 | Good | 0.158 | 20% | 7/70.00 | 3/30.00 | GLN 1, HIS 1, SER 1, THR 1, GLY 0 | LYS 1, ARG 2 |
| ISRQRLTKYT | 3.00 | 11.01 | 0.98 | 1265.48 | Good | 0.171 | 20% | 7/70.00 | 3/30.00 | GLN 1, SER 1, THR 2, GLY 0 | LYS 1, ARG 2 |
| SRQRLTKYTM | 3.00 | 11.01 | 0.98 | 1283.52 | Good | 0.114 | 20% | 7/70.00 | 3/30.00 | GLN 1, SER 1, THR 2, GLY 0 | LYS 1, ARG 2 |
| RQRLTKYTMA | 3.00 | 11.01 | 0.98 | 1267.52 | Good | 0.149 | 30% | 6/60.00 | 4/40.00 | GLN 1, THR 2, GLY 0 | LYS 1, ARG 2 |
| GERVRQALLK | 2.00 | 10.84 | 1.11 | 1169.39 | Good | 0.106 | 40% | 6/60.00 | 4/40.00 | GLN 1, GLY 1 | LYS 1, ARG 2, GLU 1 |
| RVRQALLKTV | 3.00 | 12.01 | 0.98 | 1183.46 | Good | 0.318 | 50% | 5/50.00 | 5/50.00 | GLN 1, THR 1, GLY 0 | LYS 1, ARG 2 |
| KPYIKWDLLK | 2.00 | 9.55 | 1.10 | 1303.61 | Good | 0.539 | 40% | 4/40.00 | 6/60.00 | GLY 0 | LYS 3, ASP 1 |
| RLKLFDRYFK | 3.00 | 10.29 | 1.22 | 1385.68 | Good | 0.317 | 40% | 5/50.00 | 5/50.00 | GLY 0 | LYS 2, ARG 2, ASP 1 |
| KLFDRYFKYW | 2.00 | 9.55 | 0.98 | 1465.72 | Good | 0.569 | 40% | 4/40.00 | 6/60.00 | GLY 0 | LYS 2, ARG 1, ASP 1 |
| FPFNKWGKAR | 3.00 | 11.17 | 0.98 | 1250.47 | Good | 0.327 | 40% | 5/50.00 | 5/50.00 | ASN 1, GLY 1 | LYS 2, ARG 1 |
| KWGKARLYYD | 2.00 | 9.55 | 0.98 | 1299.50 | Good | 0.242 | 30% | 5/50.00 | 5/50.00 | GLY 1 | LYS 2, ARG 1, ASP 1 |
| YAISAKNRAR | 3.00 | 11.01 | 0.86 | 1149.32 | Good | 0.004 | 40% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| AISAKNRART | 3.00 | 12.01 | 0.86 | 1087.25 | Good | -0.066 | 40% | 6/60.00 | 4/40.00 | SER 1, THR 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| KNRARTVAGV | 3.00 | 12.01 | 0.86 | 1071.25 | Good | -0.029 | 40% | 6/60.00 | 4/40.00 | THR 1, ASN 1, GLY 1 | LYS 1, ARG 2 |
| NRQFHQKLLK | 3.50 | 11.17 | 1.37 | 1311.55 | Good | 0.129 | 30% | 7/70.00 | 3/30.00 | GLN 2, HIS 1, ASN 1, GLY 0 | LYS 2, ARG 1 |
| RQFHQKLLKS | 3.50 | 11.17 | 1.37 | 1284.53 | Good | 0.185 | 30% | 7/70.00 | 3/30.00 | GLN 2, HIS 1, SER 1, GLY 0 | LYS 2, ARG 1 |
| RIMASLVLAR | 2.00 | 12.01 | 0.49 | 1129.44 | Poor | 0.621 | 70% | 3/30.00 | 7/70.00 | SER 1, GLY 0 | ARG 2 |
| RNLQHRLYEC | 1.50 | 8.57 | 0.89 | 1331.52 | Good | 0.255 | 30% | 6/60.00 | 4/40.00 | GLN 1, HIS 1, ASN 1, GLY 0 | ARG 2, GLU 1 |
| RLYECLYRNR | 2.00 | 9.36 | 0.86 | 1385.61 | Good | 0.259 | 30% | 5/50.00 | 5/50.00 | ASN 1, GLY 0 | ARG 3, GLU 1 |
| SLRCGACIRR | 3.00 | 10.43 | 0.74 | 1134.40 | Good | 0.382 | 50% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | ARG 3 |
| RCGACIRRPF | 3.00 | 10.43 | 0.74 | 1178.45 | Good | 0.467 | 50% | 4/40.00 | 6/60.00 | GLY 1 | ARG 3 |
| CGACIRRPFL | 2.00 | 9.10 | 0.49 | 1135.42 | Poor | 0.738 | 60% | 3/30.00 | 7/70.00 | GLY 1 | ARG 2 |
| GACIRRPFLC | 2.00 | 9.10 | 0.49 | 1135.42 | Poor | 0.738 | 60% | 3/30.00 | 7/70.00 | GLY 1 | ARG 2 |
| ACIRRPFLCC | 2.00 | 8.82 | 0.49 | 1181.52 | Poor | 0.892 | 70% | 2/20.00 | 8/80.00 | GLY 0 | ARG 2 |
| CIRRPFLCCK | 3.00 | 9.26 | 0.86 | 1238.61 | Good | 0.762 | 60% | 3/30.00 | 7/70.00 | GLY 0 | LYS 1, ARG 2 |
| IRRPFLCCKC | 3.00 | 9.26 | 0.86 | 1238.61 | Good | 0.762 | 60% | 3/30.00 | 7/70.00 | GLY 0 | LYS 1, ARG 2 |
| RRPFLCCKCC | 3.00 | 9.02 | 0.86 | 1228.60 | Good | 0.736 | 60% | 3/30.00 | 7/70.00 | GLY 0 | LYS 1, ARG 2 |
| MSYYCKSHKP | 2.50 | 9.17 | 0.88 | 1243.47 | Good | 0.348 | 20% | 5/50.00 | 5/50.00 | HIS 1, SER 2, GLY 0 | LYS 2 |
| ANTCTERLKL | 1.00 | 8.57 | 0.74 | 1148.35 | Good | 0.253 | 40% | 6/60.00 | 4/40.00 | THR 2, ASN 1, GLY 0 | LYS 1, ARG 1, GLU 1 |
| SWEVGKPRPP | 1.00 | 9.10 | 0.74 | 1152.32 | Good | 0.295 | 20% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | LYS 1, ARG 1, GLU 1 |
| VGKPRPPLNR | 3.00 | 12.01 | 0.86 | 1133.36 | Good | 0.147 | 20% | 5/50.00 | 5/50.00 | ASN 1, GLY 1 | LYS 1, ARG 2 |
| GKPRPPLNRN | 3.00 | 12.01 | 0.86 | 1148.33 | Good | -0.035 | 10% | 6/60.00 | 4/40.00 | ASN 2, GLY 1 | LYS 1, ARG 2 |
| KALKYLPIDK | 2.00 | 9.55 | 1.10 | 1188.48 | Good | 0.345 | 40% | 4/40.00 | 6/60.00 | GLY 0 | LYS 3, ASP 1 |
| DKCSRIIPAR | 2.00 | 9.55 | 0.86 | 1158.39 | Good | 0.235 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 0 | LYS 1, ARG 2, ASP 1 |
| KCSRIIPARA | 3.00 | 10.87 | 0.86 | 1114.38 | Good | 0.343 | 50% | 4/40.00 | 6/60.00 | SER 1, GLY 0 | LYS 1, ARG 2 |
| CSRIIPARAR | 3.00 | 11.71 | 0.74 | 1142.39 | Good | 0.341 | 50% | 4/40.00 | 6/60.00 | SER 1, GLY 0 | ARG 3 |
| SRIIPARARV | 3.00 | 12.31 | 0.74 | 1138.38 | Good | 0.309 | 50% | 4/40.00 | 6/60.00 | SER 1, GLY 0 | ARG 3 |
| RIIPARARVE | 2.00 | 11.70 | 0.86 | 1180.42 | Good | 0.249 | 50% | 4/40.00 | 6/60.00 | GLY 0 | ARG 3, GLU 1 |
| SVVNARLRAK | 3.00 | 12.01 | 0.86 | 1113.33 | Good | 0.111 | 50% | 5/50.00 | 5/50.00 | SER 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| VVNARLRAKH | 3.50 | 12.01 | 1.00 | 1163.39 | Good | 0.128 | 50% | 5/50.00 | 5/50.00 | HIS 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| VNARLRAKHY | 3.50 | 11.01 | 1.00 | 1227.44 | Good | 0.102 | 40% | 5/50.00 | 5/50.00 | HIS 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| NARLRAKHYV | 3.50 | 11.01 | 1.00 | 1227.44 | Good | 0.102 | 40% | 5/50.00 | 5/50.00 | HIS 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| PAPRTLLTKG | 2.00 | 11.01 | 0.61 | 1053.27 | Good | 0.367 | 30% | 5/50.00 | 5/50.00 | THR 2, GLY 1 | LYS 1, ARG 1 |
| APRTLLTKGT | 2.00 | 11.01 | 0.61 | 1057.26 | Good | 0.321 | 30% | 6/60.00 | 4/40.00 | THR 3, GLY 1 | LYS 1, ARG 1 |
| FNSVCRLMKT | 2.00 | 9.55 | 0.61 | 1198.48 | Good | 0.510 | 50% | 5/50.00 | 5/50.00 | SER 1, THR 1, ASN 1, GLY 0 | LYS 1, ARG 1 |
| FLGTCRRCPA | 2.00 | 9.10 | 0.49 | 1123.37 | Good | 0.584 | 50% | 4/40.00 | 6/60.00 | THR 1, GLY 1 | ARG 2 |
| DNKLKAHKDK | 2.50 | 9.55 | 1.61 | 1196.37 | Good | -0.396 | 20% | 8/80.00 | 2/20.00 | HIS 1, ASN 1, GLY 0 | LYS 4, ASP 2 |
| KLKAHKDKSA | 3.50 | 10.01 | 1.61 | 1125.34 | Good | -0.232 | 30% | 7/70.00 | 3/30.00 | HIS 1, SER 1, GLY 0 | LYS 4, ASP 1 |
| FLTRNPAWRK | 3.00 | 12.01 | 0.86 | 1288.52 | Good | 0.342 | 40% | 5/50.00 | 5/50.00 | THR 1, ASN 1, GLY 0 | LYS 1, ARG 2 |
| RNPAWRKAVF | 3.00 | 12.01 | 0.86 | 1244.47 | Good | 0.299 | 50% | 4/40.00 | 6/60.00 | ASN 1, GLY 0 | LYS 1, ARG 2 |
| GIPKDMTYRR | 2.00 | 10.00 | 0.86 | 1236.46 | Good | 0.119 | 20% | 6/60.00 | 4/40.00 | THR 1, GLY 1 | LYS 1, ARG 2, ASP 1 |
| DMTYRRLISM | 1.00 | 9.10 | 0.49 | 1285.55 | Good | 0.435 | 40% | 5/50.00 | 5/50.00 | SER 1, THR 1, GLY 0 | ARG 2, ASP 1 |
| GNPKAIKCVP | 2.00 | 9.36 | 0.73 | 1026.27 | Good | 0.373 | 40% | 4/40.00 | 6/60.00 | ASN 1, GLY 1 | LYS 2 |
| WNTFTRLQSL | 1.00 | 10.11 | 0.37 | 1265.43 | Poor | 0.609 | 40% | 6/60.00 | 4/40.00 | GLN 1, SER 1, THR 2, ASN 1, GLY 0 | ARG 1 |
| ELWAKRNIKP | 2.00 | 10.01 | 1.11 | 1254.50 | Good | 0.255 | 40% | 5/50.00 | 5/50.00 | ASN 1, GLY 0 | LYS 2, ARG 1, GLU 1 |
| LWAKRNIKPV | 3.00 | 11.17 | 0.98 | 1224.52 | Good | 0.441 | 50% | 4/40.00 | 6/60.00 | ASN 1, GLY 0 | LYS 2, ARG 1 |
| WAKRNIKPVP | 3.00 | 11.17 | 0.98 | 1208.47 | Good | 0.343 | 40% | 4/40.00 | 6/60.00 | ASN 1, GLY 0 | LYS 2, ARG 1 |
| RNIKPVPEVK | 2.00 | 10.01 | 1.11 | 1179.43 | Good | 0.145 | 30% | 5/50.00 | 5/50.00 | ASN 1, GLY 0 | LYS 2, ARG 1, GLU 1 |
| LLIGLAKRFK | 3.00 | 11.17 | 0.98 | 1158.50 | Good | 0.601 | 60% | 4/40.00 | 6/60.00 | GLY 1 | LYS 2, ARG 1 |
| GLAKRFKESP | 2.00 | 10.01 | 1.11 | 1132.33 | Good | 0.085 | 30% | 6/60.00 | 4/40.00 | SER 1, GLY 1 | LYS 2, ARG 1, GLU 1 |
| KMQRMLLEKC | 2.00 | 9.72 | 1.23 | 1279.66 | Good | 0.355 | 50% | 5/50.00 | 5/50.00 | GLN 1, GLY 0 | LYS 2, ARG 1, GLU 1 |
| VLRQWLPTGT | 1.00 | 10.11 | 0.37 | 1170.38 | Poor | 0.688 | 40% | 5/50.00 | 5/50.00 | GLN 1, THR 2, GLY 1 | ARG 1 |
| DMSKFPLKLR | 2.00 | 10.01 | 0.98 | 1234.53 | Good | 0.334 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 0 | LYS 2, ARG 1, ASP 1 |
| MSKFPLKLRG | 3.00 | 11.17 | 0.98 | 1176.49 | Good | 0.411 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | LYS 2, ARG 1 |
| SKFPLKLRGT | 3.00 | 11.17 | 0.98 | 1146.40 | Good | 0.314 | 30% | 6/60.00 | 4/40.00 | SER 1, THR 1, GLY 1 | LYS 2, ARG 1 |
| KFPLKLRGTA | 3.00 | 11.17 | 0.98 | 1130.40 | Good | 0.349 | 40% | 5/50.00 | 5/50.00 | THR 1, GLY 1 | LYS 2, ARG 1 |
| MILSLLSKGR | 2.00 | 11.01 | 0.61 | 1117.42 | Good | 0.605 | 50% | 5/50.00 | 5/50.00 | SER 2, GLY 1 | LYS 1, ARG 1 |
| LLSKGRLIIR | 3.00 | 12.01 | 0.86 | 1168.49 | Good | 0.565 | 50% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | LYS 1, ARG 2 |
| GRLIIRENNR | 2.00 | 11.70 | 0.86 | 1240.43 | Good | 0.043 | 30% | 7/70.00 | 3/30.00 | ASN 2, GLY 1 | ARG 3, GLU 1 |
| RLIIRENNRV | 2.00 | 11.70 | 0.86 | 1282.51 | Good | 0.165 | 40% | 6/60.00 | 4/40.00 | ASN 2, GLY 0 | ARG 3, GLU 1 |
| ORF6 | |||||||||||
| LIIKNLSKSL | 2.00 | 10.02 | 0.73 | 1128.42 | Good | 0.604 | 50% | 5/50.00 | 5/50.00 | SER 2, ASN 1, GLY 0 | LYS 2 |
| ORF7a | |||||||||||
| HVYQLRARSV | 2.50 | 10.84 | 0.76 | 1228.42 | Good | 0.326 | 40% | 5/50.00 | 5/50.00 | GLN 1, HIS 1, SER 1, GLY 0 | ARG 2 |
| QLRARSVSPK | 3.00 | 12.01 | 0.98 | 1141.34 | Good | 0.064 | 30% | 6/60.00 | 4/40.00 | GLN 1, SER 2, GLY 0 | LYS 1, ARG 2 |
| RARSVSPKLF | 3.00 | 12.01 | 0.98 | 1160.39 | Good | 0.265 | 40% | 5/50.00 | 5/50.00 | SER 2, GLY 0 | LYS 1, ARG 2 |
| RSVSPKLFIR | 3.00 | 12.01 | 0.86 | 1202.47 | Good | 0.414 | 40% | 5/50.00 | 5/50.00 | SER 2, GLY 0 | LYS 1, ARG 2 |
| ITLCFTLKRK | 3.00 | 10.07 | 0.98 | 1222.56 | Good | 0.606 | 50% | 5/50.00 | 5/50.00 | THR 2, GLY 0 | LYS 2, ARG 1 |
| TLCFTLKRKT | 3.00 | 10.07 | 0.98 | 1210.51 | Good | 0.452 | 40% | 6/60.00 | 4/40.00 | THR 3, GLY 0 | LYS 2, ARG 1 |
| LCFTLKRKTE | 2.00 | 9.36 | 1.11 | 1238.52 | Good | 0.362 | 40% | 6/60.00 | 4/40.00 | THR 2, GLY 0 | LYS 2, ARG 1, GLU 1 |
| ORF8 | |||||||||||
| SKWYIRVGAR | 3.00 | 11.01 | 0.86 | 1235.46 | Good | 0.349 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | LYS 1, ARG 2 |
| KWYIRVGARK | 4.00 | 11.10 | 1.22 | 1276.55 | Good | 0.254 | 40% | 5/50.00 | 5/50.00 | GLY 1 | LYS 2, ARG 2 |
| YIRVGARKSA | 3.00 | 11.01 | 0.86 | 1120.32 | Good | 0.155 | 40% | 5/50.00 | 5/50.00 | SER 1, GLY 1 | LYS 1, ARG 2 |
Evaluation of membrane-binding potential of CPPs
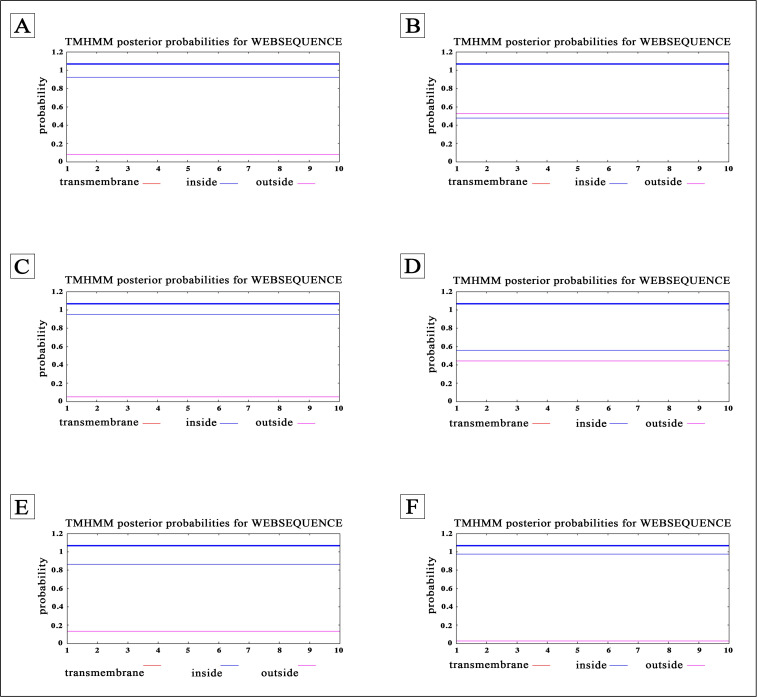
One of the principal criterions to design a potent CPP is the prediction of membrane-binding ability and cellular localization. Hence, the Boman index of each peptide was estimated using APD3 web server. The values higher than 2.48 kcal/ mol define high binding potential. For example, SSRSRNSSRN peptide derived from N-protein had the highest Boman index amongst all of the predicted CPPs (Boman Index: 7.5). Moreover, the D factor was calculated for each peptide based on net charge and μH. According to the computed D factor, CPPs can be divided into three different categories including D < 0.68 as non-lipid binding (helix/random coil), 0.68 < D < 1.34 as possible lipid-binding helix, and D > 1.34 as lipid-binding helix [46]. Additionally, the cellular localization of each CPP was evaluated by TMHMM server to determine the probability of CPPs which can enter the cell. The results of membrane-binding potential and cellular localization of CPPs were indicated in Table 3. Also, some examples of TMHMM prediction results were illustrated in Fig 1.
Table 3. Membrane-binding potential and cellular localization of CPPs.
| Epitope | Protein-binding Potential (Boman index) | Hydrophobic moment (μH) | Membrane-binding potential (D factor) | Cellular localization by TMHMM Server | Total probability of N-in by TMHMM Server |
|---|---|---|---|---|---|
| S protein | |||||
| NLTTRTQLPP | 2.49 | 0.148 | 0.469 | Inside | 0.47745 |
| RFQTLLALHR | 2.3 | 0.342 | 1.147 | Inside | 0.48730 |
| YLQPRTFLLK | 1.09 | 0.225 | 0.8724 | Outside | 0.34806 |
| SVYAWNRKRI | 3.24 | 0.224 | 1.196 | Inside | 0.89106 |
| YAWNRKRISN | 4.31 | 0.162 | 1.142 | Inside | 0.91633 |
| AWNRKRISNC | 4.17 | 0.259 | 1.234 | Inside | 0.92814 |
| WNRKRISNCV | 3.95 | 0.300 | 1.273 | Inside | 0.91952 |
| RQIAPGQTGK | 2.55 | 0.081 | 0.736 | Inside | 0.71347 |
| YNYLYRLFRK | 2.96 | 0.690 | 1.641 | Inside | 0.74845 |
| YLYRLFRKSN | 3.28 | 0.567 | 1.525 | Inside | 0.69159 |
| YRLFRKSNLK | 3.82 | 0.275 | 1.5796 | Inside | 0.75816 |
| RLFRKSNLKP | 3.81 | 0.322 | 1.623 | Inside | 0.61242 |
| RKSNLKPFER | 4.98 | 0.368 | 1.337 | Inside | 0.70344 |
| KKSTNLVKNK | 3.24 | 0.355 | 1.655 | Inside | 0.87353 |
| KSTNLVKNKC | 2.56 | 0.377 | 1.345 | Inside | 0.85159 |
| HADQLTPTWR | 2.99 | 0.468 | 0.606 | Inside | 0.56152 |
| YQTQTNSPRR | 5.62 | 0.323 | 0.964 | Inside | 0.89727 |
| TQTNSPRRAR | 6.36 | 0.219 | 1.196 | Inside | 0.92515 |
| TNSPRRARSV | 5.49 | 0.292 | 1.265 | Inside | 0.85479 |
| NSPRRARSVA | 5.05 | 0.329 | 1.300 | Inside | 0.82719 |
| PRRARSVASQ | 4.94 | 0.302 | 1.275 | Inside | 0.83300 |
| KQIYKTPPIK | 1.5 | 0.194 | 1.173 | Inside | 0.77001 |
| SQILPDPSKP | 1.67 | 0.404 | 0.381 | Outside | 0.15396 |
| RLITGRLQSL | 2.07 | 0.502 | 0.897 | Inside | 0.47579 |
| M protein | |||||
| NRNRFLYIIK | 3.1 | 0.346 | 1.316 | Inside | 0.81684 |
| RNRFLYIIKL | 1.95 | 0.326 | 1.297 | Inside | 0.66588 |
| YIIKLIFLWL | -2.91 | 0.488 | 0.790 | Outside | 0.20334 |
| KLIFLWLLWP | -2.66 | 0.304 | 0.616 | Outside | 0.07090 |
| FIASFRLFAR | 1.08 | 0.522 | 1.152 | Outside | 0.32626 |
| ASFRLFARTR | 3.62 | 0.343 | 1.313 | Inside | 0.67733 |
| SFRLFARTRS | 4.14 | 0.366 | 1.335 | Inside | 0.64273 |
| FRLFARTRSM | 3.56 | 0.350 | 1.320 | Inside | 0.67490 |
| RLFARTRSMW | 3.63 | 0.589 | 1.546 | Inside | 0.78030 |
| FARTRSMWSF | 2.67 | 0.277 | 0.921 | Inside | 0.62650 |
| HGTILTRPLL | 0.41 | 0.254 | 0.569 | Outside | 0.24888 |
| GAVILRGHLR | 1.2 | 0.323 | 0.964 | Outside | 0.42609 |
| RIAGHHLGRC | 2.43 | 0.423 | 1.059 | Inside | 0.64698 |
| YSRYRIGNYK | 3.99 | 0.141 | 1.123 | Inside | 0.89352 |
| N protein | |||||
| PQNQRNAPRI | 4.74 | 0.243 | 0.889 | Inside | 0.80935 |
| ERSGARSKQR | 6.67 | 0.143 | 1.124 | Inside | 0.89938 |
| RSGARSKQRR | 7.48 | 0.114 | 1.757 | Inside | 0.94959 |
| SGARSKQRRP | 5.99 | 0.188 | 1.497 | Inside | 0.85655 |
| GARSKQRRPQ | 6.2 | 0.185 | 1.494 | Inside | 0.89829 |
| ARSKQRRPQG | 6.2 | 0.185 | 1.494 | Inside | 0.89829 |
| RSKQRRPQGL | 5.89 | 0.280 | 1.584 | Inside | 0.83224 |
| SKQRRPQGLP | 4.4 | 0.245 | 1.221 | Inside | 0.60745 |
| KQRRPQGLPN | 4.72 | 0.292 | 1.265 | Inside | 0.69009 |
| RRPQGLPNNT | 4.53 | 0.432 | 1.067 | Inside | 0.62344 |
| QIGYYRRATR | 4.54 | 0.383 | 1.351 | Inside | 0.93350 |
| IGYYRRATRR | 5.48 | 0.445 | 1.740 | Inside | 0.95251 |
| GYYRRATRRI | 5.48 | 0.564 | 1.852 | Inside | 0.95251 |
| YYRRATRRIR | 7.07 | 0.593 | 2.209 | Inside | 0.98065 |
| YRRATRRIRG | 6.96 | 0.565 | 2.183 | Inside | 0.96774 |
| RRATRRIRGG | 6.85 | 0.478 | 2.101 | Inside | 0.94656 |
| RATRRIRGGD | 6.23 | 0.365 | 1.334 | Inside | 0.90958 |
| TRRIRGGDGK | 5.38 | 0.314 | 1.286 | Inside | 0.85699 |
| RIRGGDGKMK | 3.95 | 0.321 | 1.293 | Inside | 0.80713 |
| GKMKDLSPRW | 2.76 | 0.523 | 1.153 | Inside | 0.53414 |
| SQASSRSSSR | 5.39 | 0.171 | 0.821 | Inside | 0.70911 |
| ASSRSSSRSR | 6.33 | 0.114 | 1.097 | Inside | 0.79316 |
| SSRSSSRSRN | 7.18 | 0.180 | 1.159 | Inside | 0.81855 |
| SRSSSRSRNS | 7.18 | 0.183 | 1.162 | Inside | 0.81855 |
| RSSSRSRNSS | 7.18 | 0.179 | 1.158 | Inside | 0.81855 |
| SSSRSRNSSR | 7.18 | 0.165 | 1.145 | Inside | 0.81855 |
| SSRSRNSSRN | 7.5 | 0.192 | 1.171 | Inside | 0.86592 |
| RSRNSSRNST | 7.42 | 0.194 | 1.173 | Inside | 0.90573 |
| GSSRGTSPAR | 3.89 | 0.156 | 0.807 | Inside | 0.52191 |
| AALALLLLDR | -0.63 | 0.125 | 0.118 | Outside | 0.15140 |
| ALALLLLDRL | -0.95 | 0.288 | 0.271 | Outside | 0.09169 |
| KKSAAEASKK | 3.03 | 0.277 | 1.251 | Inside | 0.85101 |
| KSAAEASKKP | 2.48 | 0.385 | 1.023 | Inside | 0.66436 |
| SAAEASKKPR | 3.42 | 0.284 | 0.928 | Inside | 0.71820 |
| AAEASKKPRQ | 3.63 | 0.306 | 0.948 | Inside | 0.79675 |
| AEASKKPRQK | 4.36 | 0.248 | 1.224 | Inside | 0.86032 |
| EASKKPRQKR | 6.04 | 0.165 | 1.475 | Inside | 0.92101 |
| ASKKPRQKRT | 5.61 | 0.096 | 1.740 | Inside | 0.94440 |
| SKKPRQKRTA | 5.61 | 0.060 | 1.706 | Inside | 0.94440 |
| KKPRQKRTAT | 5.53 | 0.087 | 1.732 | Inside | 0.96166 |
| KPRQKRTATK | 5.53 | 0.094 | 1.738 | Inside | 0.96166 |
| PRQKRTATKA | 4.79 | 0.202 | 1.510 | Inside | 0.94133 |
| RQKRTATKAY | 4.81 | 0.311 | 1.423 | Inside | 0.96930 |
| RQGTDYKHWP | 3.88 | 0.330 | 0.641 | Inside | 0.65834 |
| FPPTEPKKDK | 3.17 | 0.088 | 0.413 | Outside | 0.35377 |
| PPTEPKKDKK | 4.02 | 0.225 | 0.872 | Inside | 0.58321 |
| PTEPKKDKKK | 4.58 | 0.236 | 1.212 | Inside | 0.80442 |
| TEPKKDKKKK | 5.13 | 0.110 | 1.527 | Inside | 0.92302 |
| EPKKDKKKKA | 4.7 | 0.147 | 1.458 | Inside | 0.90887 |
| PKKDKKKKAD | 4.89 | 0.184 | 1.493 | Inside | 0.90457 |
| KKDKKKKADE | 5.57 | 0.133 | 1.115 | Inside | 0.93664 |
| TQALPQRQKK | 3.84 | 0.308 | 0.950 | Inside | 0.86179 |
| ALPQRQKKQQ | 4.14 | 0.243 | 1.219 | Inside | 0.86346 |
| ORF3a | |||||
| SASKIITLKK | 0.94 | 0.243 | 1.219 | Inside | 0.75288 |
| ASKIITLKKR | 2.09 | 0.314 | 1.452 | Inside | 0.86807 |
| SKIITLKKRW | 2.04 | 0.492 | 1.784 | Inside | 0.86095 |
| KIITLKKRWQ | 2.25 | 0.485 | 1.777 | Inside | 0.90139 |
| IITLKKRWQL | 1.21 | 0.320 | 1.292 | Inside | 0.76749 |
| KKRWQLALSK | 2.65 | 0.335 | 1.636 | Inside | 0.79484 |
| KRWQLALSKG | 2 | 0.407 | 1.704 | Inside | 0.62646 |
| VRIIMRLWLC | 0.01 | 0.429 | 1.064 | Inside | 0.63104 |
| RIIMRLWLCW | 0.18 | 0.600 | 1.226 | Inside | 0.64902 |
| IIMRLWLCWK | -0.74 | 0.517 | 1.148 | Inside | 0.61977 |
| IMRLWLCWKC | -0.38 | 0.347 | 0.987 | Inside | 0.68839 |
| MRLWLCWKCR | 1.59 | 0.156 | 1.137 | Inside | 0.81046 |
| RLWLCWKCRS | 2.17 | 0.178 | 1.158 | Inside | 0.77912 |
| LWLCWKCRSK | 1.23 | 0.223 | 1.200 | Inside | 0.74360 |
| WLCWKCRSKN | 2.39 | 0.080 | 1.065 | Inside | 0.85315 |
| LCWKCRSKNP | 2.62 | 0.198 | 1.176 | Inside | 0.78747 |
| CWKCRSKNPL | 2.62 | 0.236 | 1.212 | Inside | 0.78747 |
| KCRSKNPLLY | 2.5 | 0.134 | 1.116 | Inside | 0.67216 |
| Orf1ab | |||||
| IKRSDARTAP | 4.15 | 0.217 | 0.864 | Inside | 0.82802 |
| KRSDARTAPH | 5.11 | 0.039 | 0.696 | Inside | 0.81819 |
| PVAYRKVLLR | 1.58 | 0.313 | 1.285 | Inside | 0.60465 |
| VAYRKVLLRK | 2.13 | 0.208 | 1.516 | Inside | 0.80830 |
| AYRKVLLRKN | 3.2 | 0.224 | 1.531 | Inside | 0.85177 |
| YRKVLLRKNG | 3.29 | 0.241 | 1.547 | Inside | 0.79673 |
| RKVLLRKNGN | 3.94 | 0.200 | 1.508 | Inside | 0.80930 |
| KVLLRKNGNK | 3 | 0.234 | 1.540 | Inside | 0.77882 |
| FEIKLAKKFD | 1.45 | 0.412 | 0.718 | Inside | 0.48733 |
| KTIQPRVEKK | 3.75 | 0.289 | 1.262 | Inside | 0.88737 |
| TIQPRVEKKK | 3.75 | 0.161 | 1.141 | Inside | 0.88737 |
| IQPRVEKKKL | 3 | 0.103 | 1.087 | Inside | 0.78283 |
| SGLKTILRKG | 1.53 | 0.531 | 1.491 | Inside | 0.54410 |
| LKTILRKGGR | 2.68 | 0.625 | 1.91 | Inside | 0.71606 |
| KTILRKGGRT | 3.43 | 0.472 | 1.765 | Inside | 0.84956 |
| GNFKVTKGKA | 1.51 | 0.141 | 1.123 | Inside | 0.68622 |
| FKVTKGKAKK | 2.05 | 0.306 | 1.938 | Inside | 0.87336 |
| KVTKGKAKKG | 2.25 | 0.248 | 1.884 | Inside | 0.88617 |
| KGKAKKGAWN | 2.1 | 0.165 | 1.475 | Inside | 0.82016 |
| GGAKLKALNL | -0.25 | 0.094 | 0.748 | Outside | 0.30336 |
| SKGLYRKCVK | 2.39 | 0.571 | 1.859 | Inside | 0.83410 |
| KGLYRKCVKS | 2.39 | 0.574 | 1.861 | Inside | 0.83410 |
| GLYRKCVKSR | 3.32 | 0.456 | 1.750 | Inside | 0.85872 |
| GLLMPLKAPK | -0.87 | 0.305 | 0.947 | Outside | 0.15005 |
| QRKQDDKKIK | 6.07 | 0.223 | 1.2 | Inside | 0.95960 |
| RKQDDKKIKA | 5.33 | 0.227 | 1.204 | Inside | 0.94962 |
| KQDDKKIKAC | 3.71 | 0.420 | 1.056 | Inside | 0.92486 |
| DITFLKKDAP | 1.64 | 0.215 | 0.202 | Inside | 0.41432 |
| MLAKALRKVP | 0.61 | 0.641 | 1.595 | Inside | 0.57064 |
| LAKALRKVPT | 1.1 | 0.661 | 1.613 | Inside | 0.61542 |
| EAKTVLKKCK | 1.95 | 0.508 | 1.469 | Inside | 0.87393 |
| AKTVLKKCKS | 1.61 | 0.525 | 1.815 | Inside | 0.87056 |
| KTVLKKCKSA | 1.61 | 0.517 | 1.808 | Inside | 0.87056 |
| KSAFYILPSI | -0.7 | 0.344 | 0.654 | Outside | 0.26827 |
| KAIVSTIQRK | 2.18 | 0.455 | 1.419 | Inside | 0.89354 |
| STIQRKYKGI | 2.68 | 0.547 | 1.506 | Inside | 0.87849 |
| TIQRKYKGIK | 2.9 | 0.533 | 1.823 | Inside | 0.93039 |
| IQRKYKGIKI | 2.15 | 0.356 | 1.656 | Inside | 0.90786 |
| GARFYFYTSK | 1.8 | 0.190 | 0.839 | Inside | 0.58982 |
| ARYMRSLKVP | 2.58 | 0.412 | 1.378 | Inside | 0.72252 |
| GIEFLKRGDK | 2.68 | 0.472 | 0.775 | Inside | 0.47568 |
| DNLKTLLSLR | 2.21 | 0.363 | 0.672 | Outside | 0.37423 |
| YMSALNHTKK | 1.94 | 0.373 | 1.012 | Inside | 0.74444 |
| SALNHTKKWK | 2.48 | 0.407 | 1.374 | Inside | 0.76830 |
| ALNHTKKWKY | 2.15 | 0.311 | 1.283 | Inside | 0.81149 |
| LNHTKKWKYP | 2.34 | 0.291 | 1.264 | Inside | 0.69869 |
| NHTKKWKYPQ | 3.38 | 0.362 | 1.331 | Inside | 0.82885 |
| HTKKWKYPQV | 2.31 | 0.217 | 1.194 | Inside | 0.78945 |
| KKPASRELKV | 3.1 | 0.317 | 1.289 | Inside | 0.71408 |
| KPASRELKVT | 2.8 | 0.356 | 0.996 | Inside | 0.66033 |
| YTPSFKKGAK | 1.7 | 0.425 | 1.391 | Inside | 0.60188 |
| PSFKKGAKLL | 0.44 | 0.488 | 1.450 | Outside | 0.28608 |
| FKKGAKLLHK | 1.12 | 0.639 | 1.923 | Inside | 0.56438 |
| KKGAKLLHKP | 1.42 | 0.544 | 1.833 | Inside | 0.55042 |
| KGAKLLHKPI | 0.37 | 0.590 | 1.546 | Inside | 0.43639 |
| WCIRCLWSTK | 0.93 | 0.223 | 0.870 | Inside | 0.79516 |
| CIRCLWSTKP | 1.17 | 0.298 | 0.941 | Inside | 0.70751 |
| ANYAKPFLNK | 1.64 | 0.408 | 1.045 | Inside | 0.53508 |
| TNIVTRCLNR | 3.3 | 0.562 | 1.190 | Inside | 0.89302 |
| CTFTRSTNSR | 4.67 | 0.140 | 0.792 | Inside | 0.87592 |
| TCMMCYKRNR | 3.74 | 0.228 | 1.205 | Inside | 0.96377 |
| MCYKRNRATR | 5.42 | 0.232 | 1.539 | Inside | 0.97415 |
| CYKRNRATRV | 5.25 | 0.084 | 1.399 | Inside | 0.97070 |
| YKRNRATRVE | 6.06 | 0.094 | 1.078 | Inside | 0.95678 |
| KRNRATRVEC | 5.92 | 0.077 | 1.062 | Inside | 0.96153 |
| RNRATRVECT | 5.62 | 0.032 | 0.690 | Inside | 0.95230 |
| RDLSLQFKRP | 4.02 | 0.454 | 1.088 | Inside | 0.49113 |
| SLQFKRPINP | 2.32 | 0.319 | 0.961 | Outside | 0.39508 |
| HNIALIWNVK | 0.05 | 0.188 | 0.507 | Inside | 0.60778 |
| LSEQLRKQIR | 4.19 | 0.686 | 1.307 | Inside | 0.75607 |
| QLRKQIRSAA | 3.64 | 0.527 | 1.487 | Inside | 0.86742 |
| LRKQIRSAAK | 3.64 | 0.621 | 1.906 | Inside | 0.89068 |
| RKQIRSAAKK | 4.68 | 0.451 | 2.075 | Inside | 0.95767 |
| KQIRSAAKKN | 3.86 | 0.413 | 1.709 | Inside | 0.93797 |
| QIRSAAKKNN | 3.97 | 0.319 | 1.291 | Inside | 0.92032 |
| AAKKNNLPFK | 1.84 | 0.198 | 1.176 | Inside | 0.63148 |
| KKNNLPFKLT | 1.96 | 0.238 | 1.214 | Inside | 0.54962 |
| NNWLKQLIKV | 0.87 | 0.728 | 1.347 | Inside | 0.65372 |
| LAYYFMRFRR | 3 | 0.469 | 1.432 | Inside | 0.71897 |
| AYYFMRFRRA | 3.31 | 0.303 | 1.276 | Inside | 0.82144 |
| YYFMRFRRAF | 3.19 | 0.478 | 1.441 | Inside | 0.72131 |
| FMRFRRAFGE | 3.75 | 0.600 | 1.226 | Inside | 0.52960 |
| MRFRRAFGEY | 4.06 | 0.504 | 1.135 | Inside | 0.70022 |
| KEMYLKLRSD | 3.29 | 0.265 | 0.580 | Inside | 0.65269 |
| YNRYLALYNK | 2.25 | 0.500 | 1.132 | Inside | 0.74756 |
| RYLALYNKYK | 2.14 | 0.392 | 1.360 | Inside | 0.80198 |
| FRKMAFPSGK | 1.83 | 0.313 | 1.285 | Inside | 0.47019 |
| TANPKTPKYK | 2.67 | 0.009 | 0.998 | Inside | 0.77069 |
| ANPKTPKYKF | 2.12 | 0.169 | 1.149 | Inside | 0.63687 |
| PKTPKYKFVR | 2.72 | 0.292 | 1.595 | Inside | 0.73005 |
| KTPKYKFVRI | 2.23 | 0.323 | 1.624 | Inside | 0.82704 |
| RWFLNRFTTT | 3.09 | 0.360 | 0.999 | Inside | 0.70898 |
| FQSAVKRTIK | 2.37 | 0.618 | 1.573 | Inside | 0.83635 |
| SEVVLKKLKK | 1.44 | 0.508 | 1.469 | Inside | 0.64932 |
| VVLKKLKKSL | 0.27 | 0.612 | 1.897 | Inside | 0.55852 |
| VLKKLKKSLN | 1.34 | 0.757 | 2.034 | Inside | 0.62971 |
| KKLKKSLNVA | 1.65 | 0.534 | 1.824 | Inside | 0.74490 |
| DAAMQRKLEK | 3.62 | 0.368 | 0.677 | Inside | 0.81844 |
| AAMQRKLEKM | 2.51 | 0.518 | 1.148 | Inside | 0.83287 |
| MQRKLEKMAD | 3.56 | 0.646 | 0.939 | Inside | 0.82292 |
| YKQARSEDKR | 6.37 | 0.233 | 0.879 | Inside | 0.92834 |
| KQARSEDKRA | 6.17 | 0.232 | 0.879 | Inside | 0.92248 |
| QARSEDKRAK | 6.17 | 0.186 | 0.835 | Inside | 0.92248 |
| MLFTMLRKLD | 0.93 | 0.562 | 0.860 | Outside | 0.39049 |
| QDLKWARFPK | 2.82 | 0.337 | 0.978 | Inside | 0.59730 |
| DLKWARFPKS | 2.61 | 0.356 | 0.996 | Inside | 0.48806 |
| LKWARFPKSD | 2.61 | 0.331 | 0.972 | Inside | 0.48806 |
| KGFCDLKGKY | 1.44 | 0.467 | 1.1 | Inside | 0.55047 |
| GVSAARLTPC | 0.6 | 0.109 | 0.432 | Inside | 0.44779 |
| GFAKFLKTNC | 0.53 | 0.530 | 1.160 | Inside | 0.51312 |
| KTNCCRFQEK | 4.2 | 0.333 | 0.974 | Inside | 0.92234 |
| PHISRQRLTK | 4.17 | 0.167 | 1.147 | Inside | 0.78638 |
| HISRQRLTKY | 4.18 | 0.190 | 1.169 | Inside | 0.87931 |
| ISRQRLTKYT | 3.97 | 0.208 | 1.186 | Inside | 0.90259 |
| SRQRLTKYTM | 4.23 | 0.231 | 1.208 | Inside | 0.90877 |
| RQRLTKYTMA | 3.71 | 0.210 | 1.188 | Inside | 0.92213 |
| GERVRQALLK | 3.11 | 0.401 | 1.038 | Inside | 0.66110 |
| RVRQALLKTV | 2.37 | 0.490 | 1.452 | Inside | 0.78888 |
| KPYIKWDLLK | 0.84 | 0.143 | 0.794 | Inside | 0.49602 |
| RLKLFDRYFK | 3.4 | 0.622 | 1.577 | Inside | 0.61889 |
| KLFDRYFKYW | 2.18 | 0.889 | 1.499 | Inside | 0.59390 |
| FPFNKWGKAR | 2.16 | 0.338 | 1.309 | Inside | 0.52642 |
| KWGKARLYYD | 2.5 | 0.177 | 0.827 | Inside | 0.72287 |
| YAISAKNRAR | 3.52 | 0.076 | 1.061 | Inside | 0.91112 |
| AISAKNRART | 3.76 | 0.037 | 1.024 | Inside | 0.91919 |
| KNRARTVAGV | 3.19 | 0.315 | 1.287 | Inside | 0.88250 |
| NRQFHQKLLK | 3.55 | 0.436 | 1.401 | Inside | 0.73407 |
| RQFHQKLLKS | 3.23 | 0.495 | 1.457 | Inside | 0.65716 |
| RIMASLVLAR | 0.84 | 0.180 | 0.829 | Inside | 0.57528 |
| RNLQHRLYEC | 4.25 | 0.519 | 0.819 | Inside | 0.77699 |
| RLYECLYRNR | 4.73 | 0.430 | 1.065 | Inside | 0.85428 |
| SLRCGACIRR | 3.3 | 0.356 | 1.326 | Inside | 0.83575 |
| RCGACIRRPF | 3.15 | 0.633 | 1.587 | Inside | 0.77458 |
| CGACIRRPFL | 1.17 | 0.422 | 1.058 | Inside | 0.52389 |
| GACIRRPFLC | 1.17 | 0.269 | 0.913 | Inside | 0.52389 |
| ACIRRPFLCC | 1.13 | 0.416 | 1.052 | Inside | 0.66539 |
| CIRRPFLCCK | 1.87 | 0.493 | 1.455 | Inside | 0.75457 |
| IRRPFLCCKC | 1.87 | 0.298 | 1.271 | Inside | 0.75457 |
| RRPFLCCKCC | 2.23 | 0.347 | 1.317 | Inside | 0.81002 |
| MSYYCKSHKP | 1.92 | 0.271 | 0.915 | Inside | 0.68047 |
| ANTCTERLKL | 2.61 | 0.348 | 0.988 | Inside | 0.76670 |
| SWEVGKPRPP | 2.33 | 0.123 | 0.446 | Outside | 0.22968 |
| VGKPRPPLNR | 3.21 | 0.403 | 1.370 | Inside | 0.41222 |
| GKPRPPLNRN | 4.28 | 0.261 | 1.236 | Inside | 0.48502 |
| KALKYLPIDK | 0.89 | 0.154 | 0.805 | Inside | 0.53339 |
| DKCSRIIPAR | 3.45 | 0.418 | 1.054 | Inside | 0.82505 |
| KCSRIIPARA | 2.4 | 0.376 | 1.344 | Inside | 0.83189 |
| CSRIIPARAR | 3.34 | 0.425 | 1.391 | Inside | 0.84740 |
| SRIIPARARV | 3.06 | 0.289 | 1.262 | Inside | 0.80072 |
| RIIPARARVE | 3.4 | 0.233 | 0.879 | Inside | 0.79867 |
| SVVNARLRAK | 2.88 | 0.144 | 1.125 | Inside | 0.81132 |
| VVNARLRAKH | 3.00 | 0.138 | 1.120 | Inside | 0.83250 |
| VNARLRAKHY | 3.42 | 0.100 | 1.084 | Inside | 0.86059 |
| NARLRAKHYV | 3.42 | 0.057 | 1.043 | Inside | 0.86059 |
| PAPRTLLTKG | 1.3 | 0.242 | 0.888 | Outside | 0.35023 |
| APRTLLTKGT | 1.55 | 0.313 | 0.955 | Inside | 0.54170 |
| FNSVCRLMKT | 1.75 | 0.603 | 1.229 | Inside | 0.71012 |
| FLGTCRRCPA | 1.92 | 0.606 | 1.232 | Inside | 0.60210 |
| DNKLKAHKDK | 4.42 | 0.269 | 0.913 | Inside | 0.80851 |
| KLKAHKDKSA | 3.04 | 0.217 | 1.194 | Inside | 0.79044 |
| FLTRNPAWRK | 3.25 | 0.256 | 1.231 | Inside | 0.70721 |
| RNPAWRKAVF | 2.9 | 0.167 | 1.147 | Inside | 0.75095 |
| GIPKDMTYRR | 3.86 | 0.181 | 0.830 | Inside | 0.81853 |
| DMTYRRLISM | 3.01 | 0.334 | 0.645 | Inside | 0.78592 |
| GNPKAIKCVP | 0.47 | 0.301 | 0.944 | Inside | 0.53539 |
| WNTFTRLQSL | 2.04 | 0.432 | 0.737 | Inside | 0.51104 |
| ELWAKRNIKP | 2.54 | 0.248 | 0.894 | Inside | 0.69594 |
| LWAKRNIKPV | 1.46 | 0.307 | 1.279 | Inside | 0.70434 |
| WAKRNIKPVP | 1.95 | 0.435 | 1.4 | Inside | 0.69474 |
| RNIKPVPEVK | 2.64 | 0.365 | 1.004 | Inside | 0.65866 |
| LLIGLAKRFK | 0.06 | 0.556 | 1.514 | Outside | 0.34306 |
| GLAKRFKESP | 2.55 | 0.567 | 1.195 | Inside | 0.41824 |
| KMQRMLLEKC | 2.25 | 0.534 | 1.164 | Inside | 0.77714 |
| VLRQWLPTGT | 0.84 | 0.442 | 0.747 | Inside | 0.37448 |
| DMSKFPLKLR | 2.29 | 0.438 | 1.073 | Outside | 0.39910 |
| MSKFPLKLRG | 1.33 | 0.425 | 1.391 | Outside | 0.33087 |
| SKFPLKLRGT | 1.82 | 0.312 | 1.284 | Outside | 0.37440 |
| KFPLKLRGTA | 1.3 | 0.337 | 1.308 | Outside | 0.41667 |
| MILSLLSKGR | 0.43 | 0.383 | 1.021 | Outside | 0.30296 |
| LLSKGRLIIR | 1.32 | 0.419 | 1.385 | Outside | 0.44682 |
| GRLIIRENNR | 4.91 | 0.336 | 0.977 | Inside | 0.81939 |
| RLIIRENNRV | 4.6 | 0.443 | 1.078 | Inside | 0.85576 |
| ORF6 | |||||
| LIIKNLSKSL | 0 | 0.601 | 1.227 | Outside | 0.38147 |
| ORF7a | |||||
| HVYQLRARSV | 2.87 | 0.205 | 0.853 | Inside | 0.75260 |
| QLRARSVSPK | 3.69 | 0.081 | 1.066 | Inside | 0.70446 |
| RARSVSPKLF | 2.84 | 0.344 | 1.314 | Inside | 0.52495 |
| RSVSPKLFIR | 2.53 | 0.158 | 1.139 | Inside | 0.51018 |
| ITLCFTLKRK | 1.21 | 0.272 | 1.246 | Inside | 0.72907 |
| TLCFTLKRKT | 1.96 | 0.280 | 1.254 | Inside | 0.78296 |
| LCFTLKRKTE | 2.38 | 0.341 | 0.981 | Inside | 0.71141 |
| ORF8 | |||||
| SKWYIRVGAR | 2.48 | 0.303 | 1.276 | Inside | 0.82389 |
| KWYIRVGARK | 2.7 | 0.208 | 1.516 | Inside | 0.89799 |
| YIRVGARKSA | 2.54 | 0.273 | 1.247 | Inside | 0.83725 |
Fig 1.
Prediction of cellular localization for CPPs using TMHMM web server: (A) EASKKPRQKR peptide containing tumor penetrating motif (RXXR); (B) GIEFLKRGDK peptide containing tumor homing motif (RGD); (C) RSGARSKQRR peptide with +5.00 net charge; (D) VVLKKLKKSL peptide with high half-life (~100 hour) in mammalian cells; (E) SSRSRNSSRN peptide with the highest Boman index (~7.5); (F) MCYKRNRATR peptide containing tumosr penetrating motif (RXXR). All of the prediction results showed the cell localization of CPPs.
Assessment of the immunogenicity
As we mentioned earlier, it is important that CPPs as a delivery system should not have any immune activity. Hence, we analyzed the immunogenicity activity of each peptide using IEDB Immunogenicity Predictor. The results were listed in Table 4.
Table 4. Evaluation of immunogenicity, toxicity, allergenicity, half-life and hemolytic potency.
| Epitope | Immunogenicity (IEDB) | Toxicity | Allergenicity (AllerTop) | Allergenicity (AllergenFP) | Half-life in E.coli | Half-life in mammalian | HemoPI |
|---|---|---|---|---|---|---|---|
| S protein | |||||||
| NLTTRTQLPP | 0.01498 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | - | - | 0.49 |
| RFQTLLALHR | 0.02623 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| YLQPRTFLLK | 0.1338 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| SVYAWNRKRI | 0.14453 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.50 |
| YAWNRKRISN | -0.02983 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| AWNRKRISNC | -0.12774 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| WNRKRISNCV | -0.21289 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| RQIAPGQTGK | 0.04829 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| YNYLYRLFRK | 0.15304 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| YLYRLFRKSN | -0.07586 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| YRLFRKSNLK | -0.21307 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.44 |
| RLFRKSNLKP | -0.42247 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| RKSNLKPFER | -0.13415 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| KKSTNLVKNK | -0.17866 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| KSTNLVKNKC | -0.29897 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| HADQLTPTWR | 0.06938 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 3.5 hour | 0.49 |
| YQTQTNSPRR | -0.20731 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| TQTNSPRRAR | -0.05055 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| TNSPRRARSV | 0.01979 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.51 |
| NSPRRARSVA | 0.0702 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.50 |
| PRRARSVASQ | -0.07931 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | - | - | 0.49 |
| KQIYKTPPIK | -0.07476 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >0 hour | ~ >20 hour | 0.49 |
| SQILPDPSKP | -0.23322 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| RLITGRLQSL | -0.0392 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| M protein | |||||||
| NRNRFLYIIK | 0.33978 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | - | - | 0.49 |
| RNRFLYIIKL | 0.2318 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.50 |
| YIIKLIFLWL | 0.17526 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| KLIFLWLLWP | 0.47552 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| FIASFRLFAR | 0.12185 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.49 |
| ASFRLFARTR | 0.29647 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| SFRLFARTRS | 0.26946 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| FRLFARTRSM | 0.18626 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.49 |
| RLFARTRSMW | -0.02793 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| FARTRSMWSF | -0.12986 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.49 |
| HGTILTRPLL | 0.2064 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 3.5 hour | 0.49 |
| GAVILRGHLR | 0.23964 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.50 |
| RIAGHHLGRC | 0.1582 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.50 |
| YSRYRIGNYK | 0.21736 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| N protein | |||||||
| PQNQRNAPRI | -0.01685 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >0 hour | ~ >20 hour | 0.49 |
| ERSGARSKQR | -0.33651 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1 hour | 0.49 |
| RSGARSKQRR | -0.33085 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| SGARSKQRRP | -0.34144 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| GARSKQRRPQ | -0.38655 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| ARSKQRRPQG | -0.36142 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| RSKQRRPQGL | -0.17416 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| SKQRRPQGLP | -0.03284 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.48 |
| KQRRPQGLPN | -0.03866 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| RRPQGLPNNT | -0.11764 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| QIGYYRRATR | 0.1585 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 0.8 hour | 0.49 |
| IGYYRRATRR | 0.19571 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 20 hour | 0.49 |
| GYYRRATRRI | 0.24984 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| YYRRATRRIR | 0.31494 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| YRRATRRIRG | 0.33565 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.48 |
| RRATRRIRGG | 0.34132 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| RATRRIRGGD | 0.3418 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| TRRIRGGDGK | 0.30454 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| RIRGGDGKMK | -0.1472 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| GKMKDLSPRW | -0.39805 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| SQASSRSSSR | -0.65648 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| ASSRSSSRSR | -0.53253 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| SSRSSSRSRN | -0.53253 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| SRSSSRSRNS | -0.4467 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| RSSSRSRNSS | -0.38427 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| SSSRSRNSSR | -0.35469 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| SSRSRNSSRN | -0.37068 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| RSRNSSRNST | -0.36531 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| GSSRGTSPAR | -0.07305 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| AALALLLLDR | 0.00733 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| ALALLLLDRL | 0.01846 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| KKSAAEASKK | -0.10752 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| KSAAEASKKP | -0.27606 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| SAAEASKKPR | -0.40103 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| AAEASKKPRQ | -0.48135 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| AEASKKPRQK | -0.60821 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| EASKKPRQKR | -0.66654 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1 hour | 0.49 |
| ASKKPRQKRT | -0.5082 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| SKKPRQKRTA | -0.2802 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| KKPRQKRTAT | -0.16998 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| KPRQKRTATK | -0.16712 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| PRQKRTATKA | -0.22281 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >0 hour | ~ >20 hour | 0.49 |
| RQKRTATKAY | -0.06462 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| RQGTDYKHWP | 0.02178 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| FPPTEPKKDK | -0.24988 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.49 |
| PPTEPKKDKK | -0.41773 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >0 hour | ~ >20 hour | 0.49 |
| PTEPKKDKKK | -0.68578 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >0 hour | ~ >20 hour | 0.49 |
| TEPKKDKKKK | -0.92 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| EPKKDKKKKA | -0.9864 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1 hour | 0.49 |
| PKKDKKKKAD | -0.82982 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >0 hour | ~ >20 hour | 0.49 |
| KKDKKKKADE | -0.78682 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| TQALPQRQKK | -0.2971 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| ALPQRQKKQQ | -0.63524 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| ORF3a | |||||||
| SASKIITLKK | -0.11032 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.56 |
| ASKIITLKKR | -0.08712 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.63 |
| SKIITLKKRW | -0.15064 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| KIITLKKRWQ | -0.16844 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| IITLKKRWQL | -0.25058 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ >20 hour | 0.50 |
| KKRWQLALSK | 0.0469 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| KRWQLALSKG | -0.29342 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| VRIIMRLWLC | 0.22654 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 100 hour | 0.49 |
| RIIMRLWLCW | 0.07375 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| IIMRLWLCWK | 0.27346 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 20 hour | 0.49 |
| IMRLWLCWKC | 0.22073 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 20 hour | 0.49 |
| MRLWLCWKCR | 0.151 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| RLWLCWKCRS | 0.00568 | Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| LWLCWKCRSK | -0.15588 | Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.48 |
| WLCWKCRSKN | -0.27401 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| LCWKCRSKNP | -0.48871 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.48 |
| CWKCRSKNPL | -0.44989 | Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.2 hour | 0.48 |
| KCRSKNPLLY | -0.39225 | Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| Orf1ab | |||||||
| IKRSDARTAP | 0.01437 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 20 hour | 0.48 |
| KRSDARTAPH | 0.1202 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| PVAYRKVLLR | -0.1276 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >0 hour | ~> 20 hour | 0.49 |
| VAYRKVLLRK | -0.10848 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >0 hour | ~ 100 hour | 0.48 |
| AYRKVLLRKN | -0.26356 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.48 |
| YRKVLLRKNG | -0.18712 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.48 |
| RKVLLRKNGN | -0.14682 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| KVLLRKNGNK | -0.15563 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| FEIKLAKKFD | -0.4631 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.47 |
| KTIQPRVEKK | -0.0364 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| TIQPRVEKKK | -0.17191 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 7.2 hour | 0.48 |
| IQPRVEKKKL | -0.32482 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 20 hour | 0.46 |
| SGLKTILRKG | -0.14596 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.52 |
| LKTILRKGGR | 0.03152 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.47 |
| KTILRKGGRT | -0.03682 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| GNFKVTKGKA | -0.4014 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.48 |
| FKVTKGKAKK | -0.42052 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.49 |
| KVTKGKAKKG | -0.65257 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| KGKAKKGAWN | -0.25629 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| GGAKLKALNL | -0.40141 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.53 |
| SKGLYRKCVK | -0.17774 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.47 |
| KGLYRKCVKS | -0.30883 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.47 |
| GLYRKCVKSR | -0.45142 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.45 |
| GLLMPLKAPK | -0.37836 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| QRKQDDKKIK | -0.4506 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 0.8 hour | 0.49 |
| RKQDDKKIKA | -0.42036 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| KQDDKKIKAC | -0.42434 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| DITFLKKDAP | -0.25182 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.49 |
| MLAKALRKVP | -0.28616 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| LAKALRKVPT | -0.16567 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.50 |
| EAKTVLKKCK | -0.41804 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1 hour | 0.49 |
| AKTVLKKCKS | -0.54116 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| KTVLKKCKSA | -0.74717 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| KSAFYILPSI | 0.14004 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| KAIVSTIQRK | 0.0192 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| STIQRKYKGI | -0.39864 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.48 |
| TIQRKYKGIK | -0.29576 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| IQRKYKGIKI | -0.39558 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 20 hour | 0.49 |
| GARFYFYTSK | 0.17762 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| ARYMRSLKVP | -0.45692 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| GIEFLKRGDK | 0.01978 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.47 |
| DNLKTLLSLR | -0.35014 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.49 |
| YMSALNHTKK | -0.09422 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| SALNHTKKWK | -0.19639 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.50 |
| ALNHTKKWKY | -0.28381 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| LNHTKKWKYP | -0.34609 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.49 |
| NHTKKWKYPQ | -0.4113 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | - | - | 0.49 |
| HTKKWKYPQV | -0.36182 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 3.5 hour | 0.49 |
| KKPASRELKV | -0.11604 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.45 |
| KPASRELKVT | -0.17419 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| YTPSFKKGAK | -0.41761 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| PSFKKGAKLL | -0.50765 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >0 hour | ~ >20 hour | 0.51 |
| FKKGAKLLHK | -0.2087 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.48 |
| KKGAKLLHKP | -0.27957 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.47 |
| KGAKLLHKPI | -0.38393 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.44 |
| WCIRCLWSTK | 0.12355 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| CIRCLWSTKP | -0.08152 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.2 hour | 0.48 |
| ANYAKPFLNK | -0.08557 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| TNIVTRCLNR | 0.10905 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| CTFTRSTNSR | -0.09922 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.2 hour | 0.49 |
| TCMMCYKRNR | -0.4529 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| MCYKRNRATR | -0.06968 | Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.48 |
| CYKRNRATRV | 0.12601 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.2 hour | 0.49 |
| YKRNRATRVE | 0.20313 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| KRNRATRVEC | 0.26794 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| RNRATRVECT | 0.23623 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| RDLSLQFKRP | -0.33523 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| SLQFKRPINP | 0.0187 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.48 |
| HNIALIWNVK | 0.42854 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 3.5 hour | 0.49 |
| LSEQLRKQIR | -0.26746 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.49 |
| QLRKQIRSAA | -0.25144 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 0.8 hour | 0.44 |
| LRKQIRSAAK | -0.10641 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.37 |
| RKQIRSAAKK | -0.07053 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| KQIRSAAKKN | -0.29889 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.50 |
| QIRSAAKKNN | -0.46225 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 0.8 hour | 0.49 |
| AAKKNNLPFK | -0.251 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| KKNNLPFKLT | -0.10849 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| NNWLKQLIKV | -0.28618 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | - | - | 0.49 |
| LAYYFMRFRR | 0.11584 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.49 |
| AYYFMRFRRA | 0.18012 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| YYFMRFRRAF | 0.14096 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| FMRFRRAFGE | 0.39083 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.49 |
| MRFRRAFGEY | 0.37588 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| KEMYLKLRSD | -0.34494 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| YNRYLALYNK | 0.02304 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| RYLALYNKYK | -0.16888 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| FRKMAFPSGK | -0.22486 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.50 |
| TANPKTPKYK | -0.38006 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| ANPKTPKYKF | -0.52572 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| PKTPKYKFVR | -0.29224 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >0 hour | ~ >30 hour | 0.49 |
| KTPKYKFVRI | -0.25892 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| RWFLNRFTTT | 0.23658 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| FQSAVKRTIK | -0.02489 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.50 |
| SEVVLKKLKK | -0.50422 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.48 |
| VVLKKLKKSL | -0.92306 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 100 hour | 0.48 |
| VLKKLKKSLN | -0.8569 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 100 hour | 0.48 |
| KKLKKSLNVA | -0.58348 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.47 |
| DAAMQRKLEK | -0.38026 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.49 |
| AAMQRKLEKM | -0.3851 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| MQRKLEKMAD | -0.44184 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| YKQARSEDKR | -0.12196 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| KQARSEDKRA | -0.1297 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| QARSEDKRAK | -0.16703 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 0.8 hour | 0.49 |
| MLFTMLRKLD | -0.2445 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| QDLKWARFPK | 0.17424 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 0.8 hour | 0.48 |
| DLKWARFPKS | 0.21623 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.49 |
| LKWARFPKSD | -0.01343 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.49 |
| KGFCDLKGKY | -0.30585 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| GVSAARLTPC | 0.09001 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.48 |
| GFAKFLKTNC | -0.27512 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| KTNCCRFQEK | 0.01249 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| PHISRQRLTK | -0.12363 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >0 hour | ~ >20 hour | 0.47 |
| HISRQRLTKY | -0.1677 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 3.5 hour | 0.48 |
| ISRQRLTKYT | -0.20778 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 20 hour | 0.49 |
| SRQRLTKYTM | -0.14196 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| RQRLTKYTMA | -0.23988 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| GERVRQALLK | 0.01693 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.48 |
| RVRQALLKTV | -0.24222 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| KPYIKWDLLK | 0.14346 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| RLKLFDRYFK | 0.16844 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.46 |
| KLFDRYFKYW | 0.03316 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| FPFNKWGKAR | -0.09005 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.49 |
| KWGKARLYYD | -0.13322 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| YAISAKNRAR | -0.23472 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| AISAKNRART | -0.11865 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| KNRARTVAGV | 0.23605 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| NRQFHQKLLK | -0.21994 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | - | - | 0.49 |
| RQFHQKLLKS | -0.39805 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| RIMASLVLAR | -0.13717 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| RNLQHRLYEC | 0.00668 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| RLYECLYRNR | 0.07267 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| SLRCGACIRR | 0.12546 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.46 |
| RCGACIRRPF | 0.21339 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| CGACIRRPFL | 0.24621 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.2 hour | 0.43 |
| GACIRRPFLC | 0.2991 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.43 |
| ACIRRPFLCC | 0.20422 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.48 |
| CIRRPFLCCK | 0.08464 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.2 hour | 0.49 |
| IRRPFLCCKC | -0.11341 | Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 20 hour | 0.49 |
| RRPFLCCKCC | -0.21335 | Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| MSYYCKSHKP | -0.52185 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| ANTCTERLKL | 0.00701 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| SWEVGKPRPP | -0.0762 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| VGKPRPPLNR | -0.06514 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 100 hour | 0.49 |
| GKPRPPLNRN | 0.04122 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| KALKYLPIDK | -0.12016 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| DKCSRIIPAR | 0.13481 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.48 |
| KCSRIIPARA | 0.3104 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| CSRIIPARAR | 0.37289 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.2 hour | 0.49 |
| SRIIPARARV | 0.3164 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| RIIPARARVE | 0.22517 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| SVVNARLRAK | 0.15149 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.48 |
| VVNARLRAKH | 0.03261 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 100 hour | 0.49 |
| VNARLRAKHY | -0.02189 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 100 hour | 0.48 |
| NARLRAKHYV | -0.08372 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | - | - | 0.48 |
| PAPRTLLTKG | -0.0282 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >0 hour | ~ >20 hour | 0.49 |
| APRTLLTKGT | -0.0914 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 4.4 hour | 0.49 |
| FNSVCRLMKT | -0.2989 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.48 |
| FLGTCRRCPA | 0.0447 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.47 |
| DNKLKAHKDK | -0.39165 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.49 |
| KLKAHKDKSA | -0.46691 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.48 |
| FLTRNPAWRK | 0.30159 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.1 hour | 0.48 |
| RNPAWRKAVF | 0.15601 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| GIPKDMTYRR | -0.30634 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| DMTYRRLISM | 0.1149 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.49 |
| GNPKAIKCVP | -0.27728 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.48 |
| WNTFTRLQSL | 0.01374 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| ELWAKRNIKP | -0.0681 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1 hour | 0.49 |
| LWAKRNIKPV | -0.2234 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.50 |
| WAKRNIKPVP | -0.08286 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.49 |
| RNIKPVPEVK | -0.04622 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| LLIGLAKRFK | 0.01368 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.52 |
| GLAKRFKESP | -0.25506 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.52 |
| KMQRMLLEKC | -0.21926 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| VLRQWLPTGT | 0.14726 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 100 hour | 0.49 |
| DMSKFPLKLR | -0.36642 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 1.1 hour | 0.49 |
| MSKFPLKLRG | -0.15592 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 30 hour | 0.52 |
| SKFPLKLRGT | -0.14092 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.53 |
| KFPLKLRGTA | -0.13556 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.52 |
| MILSLLSKGR | -0.5096 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.50 |
| LLSKGRLIIR | -0.00766 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.50 |
| GRLIIRENNR | 0.39533 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 30 hour | 0.49 |
| RLIIRENNRV | 0.34371 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1 hour | 0.49 |
| ORF6 | |||||||
| LIIKNLSKSL | -0.62529 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.49 |
| ORF7a | |||||||
| HVYQLRARSV | -0.09431 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 min | ~ 3.5 hour | 0.49 |
| QLRARSVSPK | -0.16177 | Non-Toxin | PROBABLE ALLERGEN | PROBABLE ALLERGEN | ~ >10 hour | ~ 0.8 hour | 0.47 |
| RARSVSPKLF | -0.46949 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| RSVSPKLFIR | -0.20775 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 1 hour | 0.48 |
| ITLCFTLKRK | -0.06825 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 20 hour | 0.49 |
| TLCFTLKRKT | -0.15802 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 7.2 hour | 0.49 |
| LCFTLKRKTE | -0.25434 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 5.5 hour | 0.49 |
| ORF8 | |||||||
| SKWYIRVGAR | 0.3385 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ >10 hour | ~ 1.9 hour | 0.49 |
| KWYIRVGARK | 0.31848 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE ALLERGEN | ~ 2 min | ~ 1.3 hour | 0.49 |
| YIRVGARKSA | -0.1005 | Non-Toxin | PROBABLE Non-ALLERGEN | PROBABLE Non-ALLERGEN | ~ 2 min | ~ 2.8 hour | 0.48 |
Determination of toxicity and allergenicity
The toxicity and allergenicity of each peptide were determined using diverse web servers (Table 4). In detail, most of the predicted CPPs were non-toxic. The toxic CPPs were derived from ORF3a and Orf1ab polyproteins. Furthermore, there are some differences between allergenicity prediction by AllerTop, and AllergenFP web servers. Some CPPs were determined as probable allergen by AllerTop, whereas they were identified as probable non-allergen by AllergenFP. It is rational to select CPPs which were determined as probable non-allergen by both web servers.
Estimation of hemolytic potency and half-life
It should be considered that high hydrophobicity of a peptide enhances its probability of hydrolysis in the host; therefore, the probability of hydrolysis and half-life of each peptide in E.coli and mammalian were evaluated using HemoPI and ProtLifePred web servers (Table 4). The results of hemolytic potency vary between 0 and 1 (i.e., 0 very unlikely to be hemolytic, and 1 very likely to be hemolytic). For example, seven predicted CPPs had the highest half-life in mammalian cells (~ 100 hours) which all of them were derived from Orf1ab polyprotein including VAYRKVLLRK, VVLKKLKKSL, VLKKLKKSLN, VGKPRPPLNR, VVNARLRAKH, VNARLRAKHY, and VLRQWLPTGT peptides.
Prediction of CPP structure
The 3D spatial shapes of CPPs were predicted by PEP-FOLD3 web server (Fig 2). Also, the helical wheel projection of these short peptides were obtained via Heliquest web server as indicated in Fig 3. A peptide comprising at least five adjacent hydrophobic residues (such as Leu, Ile, Ala, Val, Pro, Met, Phe, Trp, and Tyr) illustrates a hydrophobic face on a helical wheel projection [46].
Fig 2.
The 3D spatial shape of CPPs predicted by PEP-FOLD3 web server: (A) EASKKPRQKR peptide containing tumor penetrating motif (RXXR); (B) GIEFLKRGDK peptide containing tumor homing motif (RGD); (C) RSGARSKQRR peptide with +5.00 net charge; (D) VVLKKLKKSL peptide with high half-life (~100 hour) in mammalian cells; (E) SSRSRNSSRN peptide with the highest Boman index (~7.5); (F) MCYKRNRATR peptide containing tumor penetrating motif (RXXR).
Fig 3.
Helical wheel projection of the six selected CPPs by HeliQuest: These data indicated the possible amphipathic α-helical conformation of the selected CPPs: (A) EASKKPRQKR peptide containing tumor penetrating motif (RXXR); (B) GIEFLKRGDK peptide containing tumor homing motif (RGD); (C) RSGARSKQRR peptide with +5.00 net charge; (D) VVLKKLKKSL peptide with high half-life (~100 hour) in mammalian cells; (E) SSRSRNSSRN peptide with the highest Boman index (~7.5); (F) MCYKRNRATR peptide containing tumor penetrating motif (RXXR). The structural motifs were shown as hydrophobic (yellow) and cationic (blue). Arrow illustrates direction of the hydrophobic moment (μH).
Discussion
Vaccination is one of the most effective strategies for control of dangerous pathogens. A potent vaccine must stimulate strong humoral and cellular immune responses in host [56]. The vaccine efficacy relies on various factors including the selected antigen, adjuvant and delivery system [57]. Therefore, many researchers have focused on development of novel and powerful delivery systems [9,58–60]. Since the discovery of CPPs, these short peptides were considered as a significant delivery system to enter diverse types of cargoes into cells due to their high cellular uptake efficiency. Several viruses such as HIV-1, Influenza A virus subtype H5N1, Dengue virus and HSV-1 contain CPPs in their proteome [11,30,34,61].
The bioinformatics strategies take scientists one step forward in screening and evaluating CPPs. Hence, the current study was planned to screen and identify novel and potent CPPs in the proteome of SARS-CoV-2 using in silico tools. To achieve this aim, we extracted the sequences of S, M, N, E, ORF1ab, ORF3a, ORF6, ORF7a, ORF8, and ORF10 proteins and submitted to CellPPD web server. The CellPPD is a support vector machine (SVM)-based prediction approach which was established to predict highly efficient cell penetrating peptides. The CellPPD method was based on binary profile of peptides that settle the information of both composition and order of residues in peptides [40]. The output of analysis using CellPPD web server was a large number of CPPs which subjected to several web servers for further analysis such as their physicochemical properties, uptake efficiency, toxicity, allergenicity, cellular localization, tendency for binding to plasma membrane, and prediction of 3D structure. Our results showed that the proteome of SARS-CoV-2 contains a large number of cell penetrating peptides. Most of the predicted CPPs were originated from Orf1ab polyprotein. Orf1ab polyprotein forms about two thirds of the SARS-CoV-2 genome that is translated into two polypeptides such as pp1a and pp1b. Next, these two polypeptides are processed and cleaved into sixteen non-structural proteins (nsp). Non-structural proteins possess crucial functions in the replication, transcription and pathogenesis of viral RNA [29]. Despite Orf1ab polyprotein, our data indicated that no CPP was found in the E protein. This protein is responsible for virus production and maturation [28]. Herein, twenty-four CPPs were predicted in spike (S) protein, as well. Furthermore, most of the predicted CPPs in S protein are amphipathic in nature. On the other hand, most of the predicted CPPs showed high uptake efficiency using in silico approaches. The studies demonstrated that several factors affect the uptake efficiency such as the number of arginine, the existence of tryptophan and its affinity to form helical structure, and orientation of tryptophan and arginine around the helix [62–64]. In addition, it should be considered that CPPs because of their natural pore-forming propensity and high hydrophobic moment (μH) could damage or destabilize the lipid bilayers irreversibly and so they showed cytotoxic effects. Hence, minimizing μH should be performed to reduce the membrane-disturbing by CPPs [65,66]. Our data indicated that most of the CPPs predicted from the proteome of SARS-CoV-2 were not toxic and allergen, and had appropriate half-life, as well as they could bind to plasma membrane with high potential and subsequently penetrate into cells. For example, Kajiwara et al. showed that H5N1 highly pathogenic avian influenza virus (HPAIV) infects host cells by recruiting CPP activity of the C-terminal domain of HA1 protein (HA314-346) [61]. Moreover, the N-terminal tail of capsid protein (CaP) from the plant-infecting brome mosaic virus (BMV) containing the arginine-rich motif was essential for penetration through cellular membranes [67]. Thus, it is possible that CPPs found in the SARS-CoV-2 proteome possess the potency for virus penetration into host cells.
On the other hand, CPPs are not cell-specific and thus they are internalized in most of the cell types through receptor-independent approach. Hence, to determine CPPs that might be cancer-specific or might enter cancerous cells effectively, the peptide sequence should possess the tumor homing motif (RGD) and/or tumor penetrating motif (RXXR). Moreover, the peptides harboring RXXR motif at their C-terminal region could enter tumor cells through binding to neuropilin receptor which was commonly expressed at the surface of tumor cells [19]. In our study, one of the SARS-CoV-2-derived CPPs (i.e., GIEFLKRGDK) contains RGD motif. This CPP with +1.00 net charge was soluble in water, non-toxic, and its half-life was about 30 hours in mammalian. Its cellular localization was predicted using TMHMM server. Interestingly, two CPPs such as EASKKPRQKR and MCYKRNRATR peptides included RXXR motif at their C-terminal regions. In detail, EASKKPRQKR peptide had +4.00 net charge and good water solubility. This peptide was non-toxic and non-allergen with about one hour half-life in mammalian. Also, the Boman index was 6.04 for this CPP (i.e., the values higher than 2.48 kcal/mol showed high binding potential), and its cellular localization was confirmed by TMHMM web server. Moreover, MCYKRNRATR peptide had +4.00 net charge and good water solubility. But this CPP was predicted as a toxic and allergen peptide with the estimated Boman index about 5.42. Additionally, TMHMM web server predicted its localization inside the cell. Therefore, based on our data, the efficiency of GIEFLKRGDK and EASKKPRQKR peptides can be further evaluated in vitro and in vivo as a delivery system in cancer therapy.
In the present study, only CPPs with 10 residues in length were predicted. As known, CPPs contain 5–50 residues in length [11]. Thus, we can design novel CPPs with more length and higher efficiency by addition of some sequences for delivery of different cargoes. For instance, we can add a hydrophilic lysine-rich domain derived from NLS of SV40 large T-antigen (KKKRKV) and a spacer domain (WSQP) to improve the efficiency of CPPs in DNA delivery as used in other studies [33]. In this study, as an example, by merging 11 overlapped CPPs derived from N protein such as KKSAAEASKK, KSAAEASKKP, SAAEASKKPR, AAEASKKPRQ, AEASKKPRQK, EASKKPRQKR, ASKKPRQKRT, SKKPRQKRTA, KKPRQKRTAT, KPRQKRTATK, and PRQKRTATKA peptides (with net charges of +4.00 and +5.00), a novel CPP was designed with 21 residues in length (i.e., KKSAAEASKKPRQKRTATKAY). This CPP had +7.00 net charge and good water solubility. Moreover, it was non-allergen and non-toxic with immunogenicity score about -0.70123 and D factor about 2.46 which would be located into cells as predicted by TMHMM web server. Surprisingly, when the SV40 large T-antigen NLS sequence and a spacer domain were conjugated to this CPP, we had a new CPP with 31 amino acids in length (i.e., KKSAAEASKKPRQKRTATKAYWSQPKKKRKV), +12.00 net charge, and good water solubility. This peptide was non-allergen and non-toxic with immunogenicity score about -1.49065 and D factor about 4.11, which was localized into cells as predicted by TMHMM web server. Indeed, using the conjugation of NLS and spacer to the designed CPP, the net charge and the probability of cellular localization inside cells were enhanced. Our predicted and designed CPP is similar to MPG CPP (27 residues in length, and +4.00 net charge) composed of peptide derived from HIV-1 glycoprotein 41, SV40 NLS and spacer domain. The MPG peptide was reported for delivery of DNA-based vaccine both in vitro and in vivo [33,68,69].
Conclusion
In conclusion, novel and potent CPPs derived from the proteome of SARS-CoV-2 were identified using in silico methods. It is possible for relationship between these CPPs and rapid spreading the virus in host. Moreover, we designed a long and novel CPP conjugated to SV40 NLS and spacer domain that had high binding ability to membrane and localization inside cells. The designed CPP was similar to MPG CPP. This CPP can be further evaluated for DNA delivery in vitro and in vivo in future. Generally, the predicted and designed CPPs derived from the proteome of SARS-CoV-2 with different properties can be applied to deliver different cargoes in vaccine and drug development.
Supporting information
(TIF)
Data Availability
All relevant data are within the manuscript.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Florindo HF , Kleiner R, Vaskovich-Koubi D, Acúrcio RC, Carreira B, Yeini E, et al. Immune-mediated approaches against COVID-19. Nature Nanotechnology. 2020; 15: 630–645. 10.1038/s41565-020-0732-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zareba G. A new combination vaccine for measles, mumps, rubella and varicella. Drugs Today. 2006; 42: 321–329. 10.1358/dot.2006.42.5.973586 [DOI] [PubMed] [Google Scholar]
- 3.Kardani K, Basimi P, Fekri M, Bolhassani A. Antiviral therapy for the sexually transmitted viruses: recent updates on vaccine development. Expert Rev. Clin. Pharmacol. 2020; 13 (9): 1001–1046. [DOI] [PubMed] [Google Scholar]
- 4.Garg A, Dewangan HK. Nanoparticles as adjuvants in vaccine delivery. Crit. Rev. Ther. Drug. 2020; 37 (2): 183–204. 10.1615/CritRevTherDrugCarrierSyst.2020033273 [DOI] [PubMed] [Google Scholar]
- 5.Nevagi RJ, Skwarczynski M, Toth I. Polymers for subunit vaccine delivery. Eur. Polym. J. 2019; 114: 397–410. [Google Scholar]
- 6.Şenel S, Yüksel S. Chitosan-based particulate systems for drug and vaccine delivery in the treatment and prevention of neglected tropical diseases. Drug. Deliv. Transl. Res. 2020; 10: 1644–1674. 10.1007/s13346-020-00806-4 [DOI] [PubMed] [Google Scholar]
- 7.Yu R, Mai Y, Zhao Y, Hou Y, Liu Y, Yang J. Targeting strategies of liposomal subunit vaccine delivery systems to improve vaccine efficacy. J. Drug Target. 2019; 27: 780–789. 10.1080/1061186X.2018.1547734 [DOI] [PubMed] [Google Scholar]
- 8.Du X, Wang J, Zhou Q, Zhang L, Wang S, Zhang Z, et al. Advanced physical techniques for gene delivery based on membrane perforation. Drug Deliv. 2018; 25: 1516–1525. 10.1080/10717544.2018.1480674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bolhassani A, Jafarzade BS, Mardani G. In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides. 2017; 87: 50–63. 10.1016/j.peptides.2016.11.011 [DOI] [PubMed] [Google Scholar]
- 10.Shahbazi S, Bolhassani A. Comparison of six cell penetrating peptides with different properties for in vitro and in vivo delivery of HPV16 E7 antigen in therapeutic vaccines. Int. Immunopharmacol. 2018; 62: 170–180. 10.1016/j.intimp.2018.07.006 [DOI] [PubMed] [Google Scholar]
- 11.Kardani K, Milani A, Shabani SH, Bolhassani A. Cell penetrating peptides: the potent multi-cargo intracellular carriers. Expert Opin. Drug Deliv. 2019; 16: 1227–1258. 10.1080/17425247.2019.1676720 [DOI] [PubMed] [Google Scholar]
- 12.Hoffmann K, Milech N, Juraja SM, Cunningham PT, Stone SR, Francis RW, et al. A platform for discovery of functional cell-penetrating peptides for efficient multi-cargo intracellular delivery. Sci. Rep. 2018; 8: 1–6. 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xia H, Gao X, Gu G, Liu Z, Hu Q, Tu Y, et al. Penetratin-functionalized PEG-PLA nanoparticles for brain drug delivery. Int. J. Pharm. 2012; 436: 840–850. 10.1016/j.ijpharm.2012.07.029 [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Luo Y, Shibu MA, Toth I, Skwarczynskia M. Cell-penetrating peptides: Efficient vectors for vaccine delivery. Curr. Drug Deliv. 2019; 16: 430–443. 10.2174/1567201816666190123120915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milletti F. Cell-penetrating peptides: classes, origin, and current landscape. Drug Discov. Today. 2012; 17: 850–860. 10.1016/j.drudis.2012.03.002 [DOI] [PubMed] [Google Scholar]
- 16.Maiolo JR, Ferrer M, Ottinger EA. Effects of cargo molecules on the cellular uptake of arginine-rich cell-penetrating peptides. Biochim. Biophys. Acta Biomembr. 2005; 1712: 161–172. 10.1016/j.bbamem.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 17.Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv. Drug Deliv. Rev. 2005; 57: 547–558. 10.1016/j.addr.2004.10.009 [DOI] [PubMed] [Google Scholar]
- 18.Madani F, Lindberg S, Langel Ü, Futaki S, Gräslund A. Mechanisms of cellular uptake of cell-penetrating peptides. J. Biophys. 2011; 2011: 1–10. 10.1155/2011/414729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautam A, Sharma M, Vir P, Chaudhary K, Kapoor P, Kumar R, et al. Identification and characterization of novel protein-derived arginine-rich cell-penetrating peptides. Eur. J. Pharm. Biopharm. 2015; 89: 93–106. 10.1016/j.ejpb.2014.11.020 [DOI] [PubMed] [Google Scholar]
- 20.Kardani K, Bolhassani A. CPPsite 2.0: An available database of experimentally validated cell-penetrating peptides predicting their secondary and tertiary structures. J. Mol. Biol. 2020; 166703 10.1016/j.jmb.2020.11.002 [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Cui Y, Xiong Z, Nasiri A, Zhang A, Hu J. DeepSeqPan, a novel deep convolutional neural network model for pan-specific class I HLA-peptide binding affinity prediction. Sci. Rep. 2019; 9: 794 10.1038/s41598-018-37214-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang J, Fu J, Wang Y, Li B, Li Y, Yang Q, et al. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Brief Bioinformatics. 2020; 21: 621–636. 10.1093/bib/bby127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, Niu M, Zou Q. ELM-MHC: an improved MHC identification method with extreme learning machine algorithm. J. Proteome Res. 2019; 18: 1392–1401. 10.1021/acs.jproteome.9b00012 [DOI] [PubMed] [Google Scholar]
- 24.Yin J, Sun W, Li F, Hong J, Li X, Zhou Y, et al. VARIDT 1.0: variability of drug transporter database. Nucleic Acids Res. 2020; 48: D1042–D1050. 10.1093/nar/gkz779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Chu C, Huang T, Kong X, Cai YD. Prediction and analysis of cell-penetrating peptides using pseudo-amino acid composition and random forest models. Amino Acids. 2015; 47: 1485–1493. 10.1007/s00726-015-1974-5 [DOI] [PubMed] [Google Scholar]
- 26.Tang H, Su ZD, Wei HH, Chen W, Lin H. Prediction of cell-penetrating peptides with feature selection techniques. Biochem. Biophys. Res. Commun. 2016; 477: 150–154. 10.1016/j.bbrc.2016.06.035 [DOI] [PubMed] [Google Scholar]
- 27.Liu B, Chen J, Guo M, Wang X. Protein remote homology detection and fold recognition based on Sequence-Order Frequency Matrix. TCBB. 2017; 16: 292–300. 10.1109/TCBB.2017.2765331 [DOI] [PubMed] [Google Scholar]
- 28.Kardani K, Bolhassani A. Vaccine development against SARS-CoV-2: From virology to vaccine clinical trials. Coronaviruses. 2020; 1: 1–13. [Google Scholar]
- 29.Wabalo EK, Dubiwak AD, Gizaw TS, Kotu UG. Role of structural and functional proteins of SARS-COV-2. GSC. Biol. Pharm. Sci. 2020; 12: 117–129. [Google Scholar]
- 30.Freire JM, Veiga AS, Rego de Figueiredo I, de la Torre BG, Santos NC, Andreu D, et al. Nucleic acid delivery by cell penetrating peptides derived from dengue virus capsid protein: design and mechanism of action. FEBS. J. 2014; 281: 191–215. 10.1111/febs.12587 [DOI] [PubMed] [Google Scholar]
- 31.Rhee M, Davis P. Mechanism of uptake of C105Y, a novel cell-penetrating peptide. J. Biol. Chem. 2006; 281: 1233–1240. 10.1074/jbc.M509813200 [DOI] [PubMed] [Google Scholar]
- 32.Morris MC, Depollier J, Mery J, Heitz F, Divita G. A peptide carrier for the delivery of biologically active proteins into mammalian cells. Nat. Biotechnol. 2001; 19: 1173–1176. 10.1038/nbt1201-1173 [DOI] [PubMed] [Google Scholar]
- 33.Kadkhodayan S, Bolhassani A, Sadat SM, Irani S, Fotouhi F. The efficiency of Tat cell penetrating peptide for intracellular uptake of HIV-1 Nef expressed in E. coli and mammalian cell. Curr. Drug Deliv. 2017; 14(4): 536–542. 10.2174/1567201813666161006114448 [DOI] [PubMed] [Google Scholar]
- 34.Elliott G, O’Hare P. Intercellular trafficking and protein delivery by a herpes virus structural protein. Cell. 1997; 88: 223–233. 10.1016/s0092-8674(00)81843-7 [DOI] [PubMed] [Google Scholar]
- 35.Chakraborty H, Bhattacharjya S. Mechanistic insights of host cell fusion of SARS-CoV-1 and SARS-CoV-2 from atomic resolution structure and membrane dynamics. Biophysical Chemistry. 2020; 106438 10.1016/j.bpc.2020.106438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahajan M, Chatterjee D, Bhuvaneswari K, Pillay S, Bhattacharjya S. NMR structure and localization of a large fragment of the SARS-CoV fusion protein: Implications in viral cell fusion. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2018; 1860: 407–415. 10.1016/j.bbamem.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahajan M, Bhattacharjya S. NMR structures and localization of the potential fusion peptides and the pre-transmembrane region of SARS-CoV: Implications in membrane fusion. Biochimica et Biophysica Acta (BBA)-Biomembranes. 2015; 1848: 721–730. 10.1016/j.bbamem.2014.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar S, Maurya VK, Prasad AK, Bhatt MLB, Saxena SK. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV). Virus Disease. 2020; 31: 13–21. 10.1007/s13337-020-00571-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020; 579: 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gautam A, Chaudhary K, Kumar R, Sharma A, Kapoor P, Tyagi A, et al. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013; 11: 74 10.1186/1479-5876-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gautam A, Chaudhary K, Kumar R, Raghava GP. Computer-aided virtual screening and designing of cell-penetrating peptides In Cell-Penetrating Peptides 2015; 59–69); Humana Press, New York. [DOI] [PubMed] [Google Scholar]
- 42.Tang H, Su ZD, Wei HH, Chen W, Lin H. Prediction of cell-penetrating peptides with feature selection techniques. Biochem. Biophys. Res. Commun. 2016; 477: 150–154. 10.1016/j.bbrc.2016.06.035 [DOI] [PubMed] [Google Scholar]
- 43.Wei L, Xing P, Su R, Shi G, Ma ZS, Zou Q. CPPred-RF: a sequence-based predictor for identifying cell-penetrating peptides and their uptake efficiency. J. Proteome Res. 2017; 16: 2044–2053. 10.1021/acs.jproteome.7b00019 [DOI] [PubMed] [Google Scholar]
- 44.Manavalan B, Subramaniyam S, Shin TH, Kim MO, Lee G. Machine-learning-based prediction of cell-penetrating peptides and their uptake efficiency with improved accuracy. J. Proteome Res. 2018; 17: 2715–2726. 10.1021/acs.jproteome.8b00148 [DOI] [PubMed] [Google Scholar]
- 45.Boman HG. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 2003; 254: 197–215. 10.1046/j.1365-2796.2003.01228.x [DOI] [PubMed] [Google Scholar]
- 46.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific α-helical properties. Bioinformatics. 2008; 24: 2101–2102. 10.1093/bioinformatics/btn392 [DOI] [PubMed] [Google Scholar]
- 47.Sonnhammer EL, Von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. InIsmb 1998; 6: 175–182. [PubMed] [Google Scholar]
- 48.Shankar G, Arkin S, Cocea L, Devanarayan V, Kirshner S, Kromminga A, et al. Assessment and reporting of the clinical immunogenicity of therapeutic proteins and peptides-harmonized terminology and tactical recommendations. AAPS. J. 2014; 16: 658–673. 10.1208/s12248-014-9599-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuriakose A, Chirmule N, Nair P. Immunogenicity of biotherapeutics: causes and association with posttranslational modifications. J. Immunol. Res. 2016; 2016: 1–18. 10.1155/2016/1298473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, et al. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput. Biol. 2013; 9: e1003266 10.1371/journal.pcbi.1003266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R, Raghava GP. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013; 8: e73957 10.1371/journal.pone.0073957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimitrov I, Bangov I, Flower DR, Doytchinova I. AllerTOP v. 2-a server for in silico prediction of allergens. J. Mol. Model. 2014; 20: 2278 10.1007/s00894-014-2278-5 [DOI] [PubMed] [Google Scholar]
- 53.Dimitrov I, Naneva L, Doytchinova I, Bangov I. AllergenFP: allergenicity prediction by descriptor fingerprints. Bioinformatics. 2014; 30: 846–851. 10.1093/bioinformatics/btt619 [DOI] [PubMed] [Google Scholar]
- 54.Chaudhary K, Kumar R, Singh S, Tuknait A, Gautam A, Mathur D, et al. A web server and mobile app for computing hemolytic potency of peptides. Sci. Rep. 2016; 6: 1–3. 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamiable A, Thévenet P, Rey J, Vavrusa M, Derreumaux P, Tufféry P. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 2016; 44: W449–W454. 10.1093/nar/gkw329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nabel GJ. Designing tomorrow’s vaccines. N. Engl. J. Med. 2013; 368: 551–560. 10.1056/NEJMra1204186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target Ther. 2020; 5: 1–4. 10.1038/s41392-019-0089-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dhakal S, Renukaradhya GJ. Nanoparticle-based vaccine development and evaluation against viral infections in pigs. Vet Res. 2019; 50: 90 10.1186/s13567-019-0712-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wallis J, Katti P, Martin AM, Hills T, Seymour LW, Shenton DP, et al. A liposome-based cancer vaccine for a rapid and high-titre anti-ErbB-2 antibody response. Eur. J. Pharm. Sci. 2020; 152: 105456. [DOI] [PubMed] [Google Scholar]
- 60.Zhang M, Hong Y, Chen W, Wang C. Polymers for DNA vaccine delivery. ACS Biomater. Sci. Eng. 2017; 3: 108–125. 10.1021/acsbiomaterials.6b00418 [DOI] [PubMed] [Google Scholar]
- 61.Kajiwara N, Nomura N, Ukaji M, Yamamoto N, Kohara M, Yasui F, et al. Cell-penetrating peptide-mediated cell entry of H5N1 highly pathogenic avian influenza virus. Scientific Reports. 2020; 10: 1–3. 10.1038/s41598-019-56847-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rydberg HA, Matson M, Amand HL, Esbjorner EK, Nordén B. Effects of tryptophan content and backbone spacing on the uptake efficiency of cell-penetrating peptides. Biochemistry. 2012; 51: 5531–5539. 10.1021/bi300454k [DOI] [PubMed] [Google Scholar]
- 63.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The design, synthesis, and evaluation of molecules that enable or enhance cellular uptake: peptoid molecular transporters. Proc. Natl. Acad. Sci. 2000; 97: 13003–13008. 10.1073/pnas.97.24.13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caesar CE, Esbjörner EK, Lincoln P, Nordén B. Membrane interactions of cell-penetrating peptides probed by tryptophan fluorescence and dichroism techniques: correlations of structure to cellular uptake. Biochemistry. 2006; 45: 7682–7692. 10.1021/bi052095t [DOI] [PubMed] [Google Scholar]
- 65.Hällbrink M, Florén A, Elmquist A, Pooga M, Bartfai T, Langel Ü. Cargo delivery kinetics of cell-penetrating peptides. Biochim. Biophys. Acta Biomembr. 2001; 1515: 101–109. 10.1016/s0005-2736(01)00398-4 [DOI] [PubMed] [Google Scholar]
- 66.Räägel H, Pooga M. Peptide and protein delivery with cell-penetrating peptides In Peptide and Protein Delivery 2011; 221–246; Academic Press. [Google Scholar]
- 67.Qi X, Droste T, Kao CC. Cell-penetrating peptides derived from viral capsid proteins. Molecular Plant-Microbe Interactions 2011; 24: 25–36. 10.1094/MPMI-07-10-0147 [DOI] [PubMed] [Google Scholar]
- 68.Habault J, Poyet JL. Recent advances in cell penetrating peptide-based anticancer therapies. Molecules. 2019; 24: 927 10.3390/molecules24050927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwon SJ, Han K, Jung S, Lee JE, Park S, Cheon YP, Lim HJ. Transduction of the MPG-tagged fusion protein into mammalian cells and oocytes depends on amiloride-sensitive endocytic pathway. BMC Biotechnology. 2009; 9: 73 10.1186/1472-6750-9-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Data Availability Statement
All relevant data are within the manuscript.