Figure 4. Lineage tracing with the Sleeping Beauty (SB) mouse model.
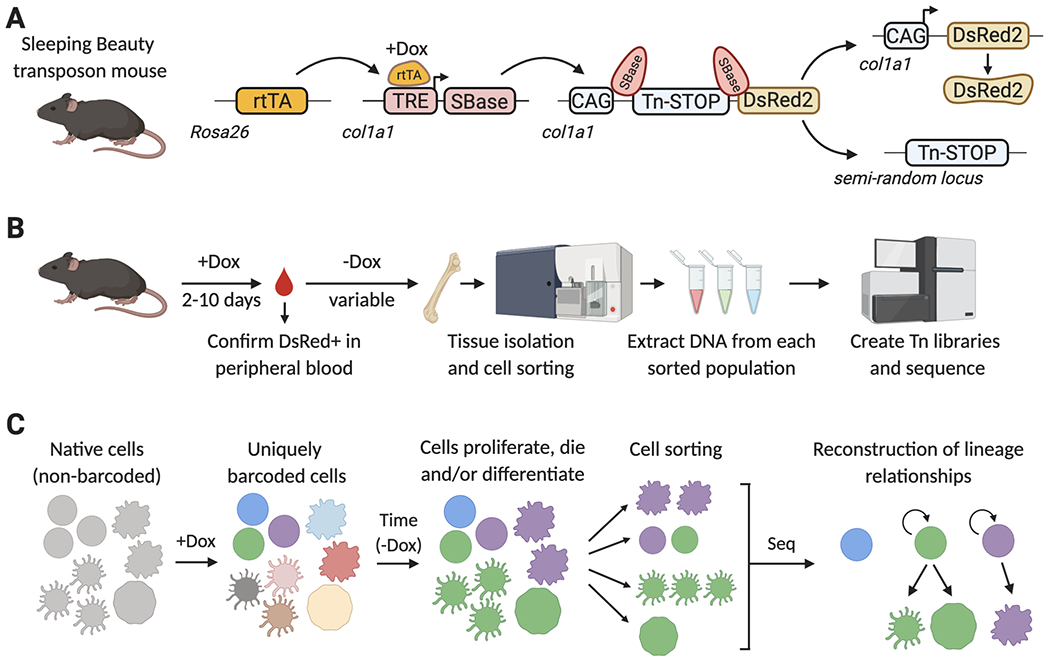
A) SB mouse model. A Dox-inducible transcriptional activator (rtTAM2), an rtTA- and Dox-regulated hyperactive SB transposase (TRE-SBase), and a single-copy SB-dependent transposable element (Tn), are combined in order to allow the inducible mobilization of the Tn element into a semi-random locus in the genome. The Tn element carries a transcriptional STOP cassette, which prevents DsRed2 expression in non-barcoded cells. Upon SBase-mediated Tn excision, DsRed2 is expressed to help avoid processing of non-barcoded cells. B) Summary of the SB-barcoding protocol. Mice are treated with Dox for up to 10 days, and expression of DsRed is confirmed in peripheral blood by flow cytometry. Dox is removed, to prevent further SBase expression and Tn mobilization. During the period of time without Dox, barcoded stem and progenitor cells will divide and differentiate, propagating these barcodes to their progeny. Tissues are then isolated and labeled with fluorescently-tagged antibodies to sort distinct cell populations by FACS. Genomic DNA is isolated from each population, and used to generate Tn-insertion libraries, which specifically amplify the Tn-flanking sequences and add adapters for next-generation sequencing. C) Reconstructing lineages with the SB mouse model. Before Dox addition, each cell in the SB barcoding mouse carries the Tn element in the same col1a1 locus. Upon Dox administration, Tn elements are mobilized and each cell will now have a unique single Tn integration site in a different locus of their genome. Upon Dox withdrawal, this integration site remains stable and establishes a tag or barcode that can be inherited by the cell progeny as HSPCs divide and differentiate into one or multiple cell types. Lists of transposon insertion sites are determined for each FACS-isolated population by next-generation sequencing, and then these are mathematically analyzed to establish lineage relationships between populations.
