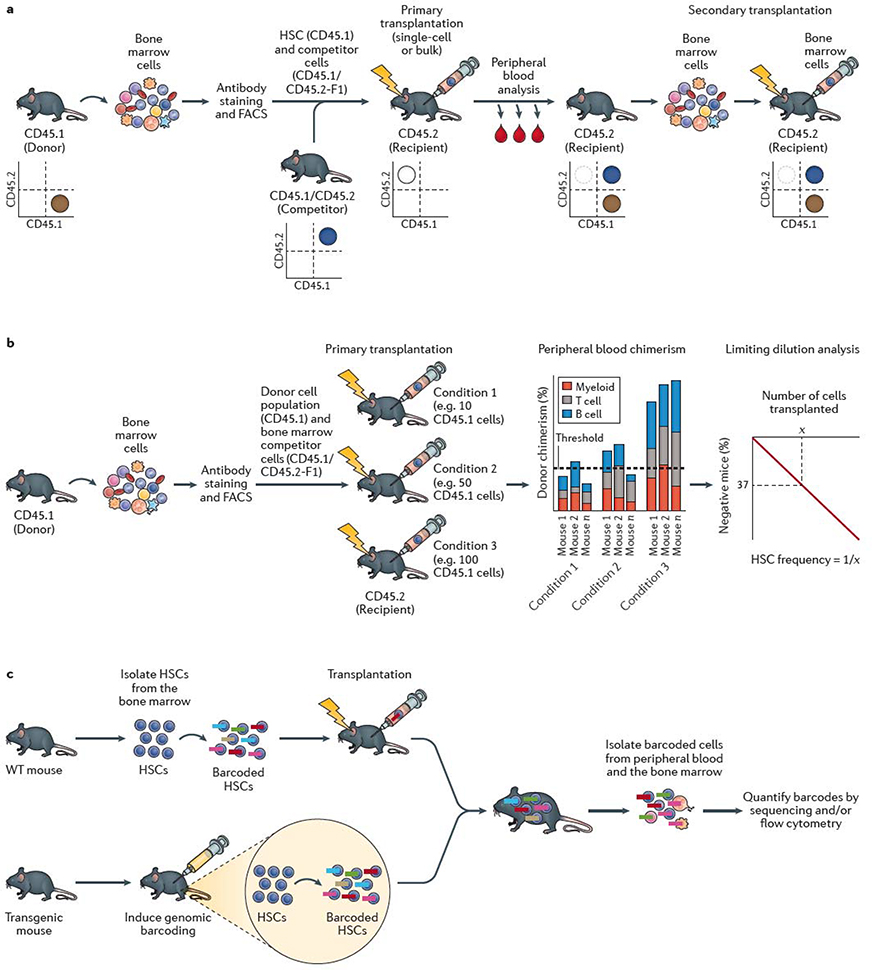
Figure 1 |. HSC self-renewal assays.
a | The standard mouse haematopoietic stem cell (HSC) transplantation assay uses the congenic CD45.1/CD45.2 system. Bone marrow cells are collected from a donor mouse bearing the CD45.1 allele from which HSCs are isolated using fluorescence-actiivated cell sorting (FACS). HSCs are mixed with competitor (also termed helper) bone marrow cells from CD45.1 × CD45.2 F1 mice and transplanted into a primary recipient mouse bearing the CD45.2 allele. Donor cell reconstitution kinetics can be determined within the peripheral blood and bone marrow using monoclonal antibodies and flow cytometric analysis. Flow cytometric quantification of donor chimerism within the peripheral blood is usually performed regularly over 16–24 weeks, and bone marrow analyzed at the study endpoint. As annotated in the representative flow cytometry plots, donor hematopoietic cells are CD45.1+, competitor hematopoietic cells are CD45.1+CD45.2+, while recipient hematopoietic cells are CD45.2+ (although irradation leads to loss of endogenous CD45.2+ haematopoietic cells in the transplant recipient). Bone marrow cells from the primary recipient mouse are transplanted into a secondary recipient mouse bearing the CD45.2 allele to confirm HSC self-renewal activity. Depending on the experimental design, CD45.1 recipient mice and CD45.2 donor mice may also be used. b | Limiting dilution analysis (LDA) can be used to estimate the frequency of HSCs within a cell population by transplantion of varying doses of cells into multiple mice. Presence or absence of an HSC(s) within the donor population is determined by assessing long-term peripheral blood reconstitution and LDA performed based on the number of positive versus negative mice as a function of the donor cell dose. Various reconstitution thresholds have been used in the field, but it is typically set at 1% donor peripheral blood chimerism. Based on Poisson statistics, the number of cells (X) that result in 37% negative mice is equal to the 1/HSC frequency. c | HSC self-renewal can be tracked using barcoding (or other genetic labelling) technologies. Barcoding can be performed ex vivo, by isolating HSCs from the bone marrow and labeling them in vitro with genetic barcodes (which can be applied using various methods, including transduction with a lentiviral library) and transplanted into recipient mice. Barcoding can also be performed directly in vivo, usually via inducing activity of a recombinase-based or transposon-based ‘shuffling’ of genetic sequences to generate unique genomic sequences in each HSC. The output of individual HSCs can be determined by quantifying the barcode abundance within peripheral blood and/or bone marrow cell populations. In vivo lineage tracing technologies can be used to track HSC self-renewal in the native bone marrow without the requirement for transplantation.