Fig. 1. Kinetic profiling of dCas9 binding by massively parallel filter binding.
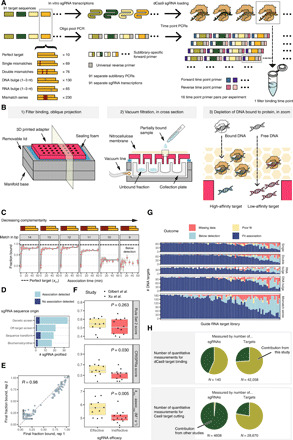
(A) Experimental overview: Cas9 targets designed to 90 sgRNAs are synthesized on an oligonucleotide array. Ninety-one distinct sublibrary primers amplify each sgRNA’s targets separately. Each library is barcoded with forward and reverse barcodes in a second PCR. Filter binding is performed, and all time points and sublibraries are pooled for sequencing. (B) Three views: (1) A plate adaptor sits on a 96-well plate vacuum manifold; (2) sample is passed through the nitrocellulose membrane and collected in a deep 96-well plate; (3) sample passes through the nitrocellulose, retaining bound DNA in the membrane. (C) Example association curves for λ1 off-target sequences. Yellow bars signify 90% confidence intervals. Dashed line signifies perfect target final fraction bound. (D) Summary of sgRNAs included in study. (E) Reproducibility of λ1 targets’ final fraction bound across two sublibraries and sgRNA preparations. Bold gray lines (lower left) denote limits of detection. (F) Comparison of “Rule Set 2” scores, CRISPRia, and on-rates in discriminating effective versus ineffective sgRNAs, as determined by genetic screening. (G) Summary of association curves for all studied sgRNA sublibraries. (H) Pie chart demonstrating scale of presented data relative to other published studies of Cas9 off-target activity.
