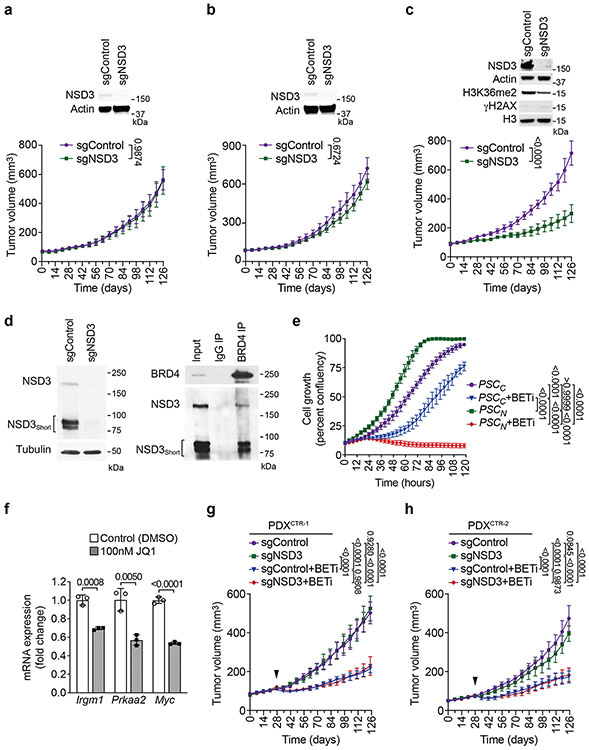
Extended Figure 9. NSD3-dependency renders PDXs from LUSC patients therapeutically vulnerable to BETi.
a-c, Tumor volume quantification of PDXCTR-1 (a), PDXCTR-2 (b) and PDXAMP-2 (c) growing in immunocompromised mice (n = 5 mice, for each treatment group). Western blots with the indicated antibodies of lysates from the PDX ± sgNSD3 is shown. Actin is shown as a loading control. d, BRD4 interacts with NSD3 in PSCN cells. Left panel: Western blot of whole cell lysates from PSCN cells ± sgNSD3 to show the relative position of NSD3 and NSD3Short in PSCN cells. Tubulin used as a loading control. Right panel: BRD4 interacts with both NSD3 and NSD3Short isoforms in PSCN cells. Co-immunoprecipitation (IP) experiments in PSCN cells with indicated antibodies for IPs and western analyses. Input, PSCN cell nuclear extract. e, Proliferation assay of PSCC and PSCN treated with vehicle control or 20nM AZD5153 (BETi). Data represent mean ± s.e.m. of three technical replicates in two independent experiments. f, Treatment of PSCN cells with the BETi JQ1 inhibits expression of the indicated NSD3 target genes. Real-time (RT) qPCR analysis of the indicated mRNAs from PSCN cells treated with DMSO or 100nM of JQ1. The RT-qPCR data for each target gene were normalized to Actb and presented as fold change relative to DMSO treated sample. Error bars represent mean ± s.d. from three independent experiments. P values were determined by two-tailed unpaired t-test. g-h, Tumor volume quantification of PDXCTR-1 (g) and PDXCTR-2 (h) treated with BETi or vehicle (control). Arrowhead indicates start of the treatment. P values were determined by two-tailed unpaired t-test. (a-c, f) or two-way ANOVA with Tukey’s post hoc test (e, g-h). Data are represented as mean ± s.e.m. (a-c, e, f-h).