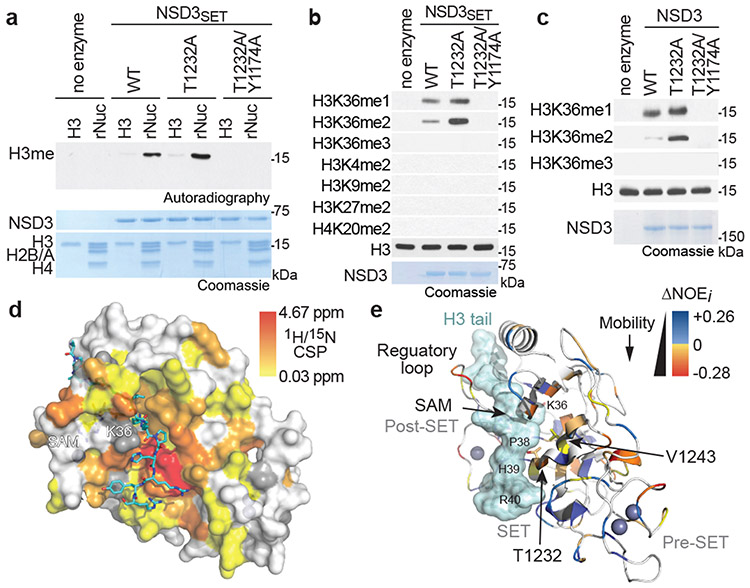
Figure 2. Molecular basis of increased H3K36me2 catalysis by NSD3T1232A.
a, In vitro methylation reactions of wild-type (WT) or mutant NSD3SET using recombinant free histone H3 or nucleosome (rNuc) and 3H-SAM as substrates. No enzyme is a negative control. Top panel, autoradiography; bottom panel, Coomassie blue staining. b, Methylation assays on rNuc as in (a) with non-radiolabeled SAM. Westerns of reaction products shown with the indicated antibodies. H3 is shown as loading control. c, Methylation assays as in (b) with recombinant full-length wild-type or mutant NSD3. Data in a-c are representative of three or more biologically independent experiments with similar results. d-e, T1232A substitution induces widespread mobility changes on the NSD3 catalytic domain. d, Amide CSPs between NSD3SET-T1232A and NSD3SET is shown mapped onto the surface representation of the NSD3SET structure (PDB: 6CEN) with docked H3.1 residues A29 to R42 (stick representation). Gray: prolines and residues missing amide assignments. e, Changes in heteronuclear {1H}-15N nuclear Overhauser effect (NOE) values plotted on the structure of NSD3 as in (d) in ribbon representation. T1232 and V1243 residues are indicated. Uncolored residues: undetectable NOE changes.