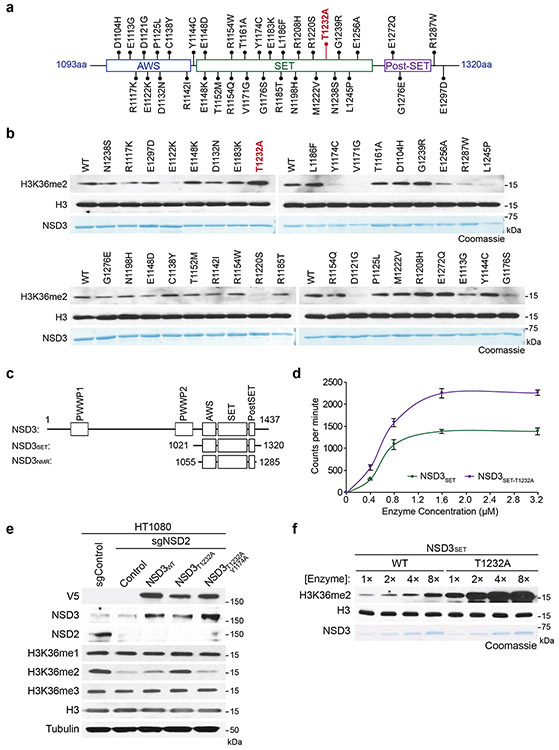
Extended Figure 3. Identification of the cancer-associated NSD3T1232A variant as a gain of function variant.
a, Schematic diagram of the distribution of the different NSD3 candidate mutants tested in (b). b, In vitro methylation assay as in Figure 2b on rNuc with recombinant wild-type NSD3SET or the indicated mutants (1-35) with non-radiolabeled SAM (the T1232A mutant is indicated in red). For each screening panel set, the top of the panel shows the Western blot and the bottom panel, Coomassie blue stain of the NSD3 proteins. c, Schematic diagram of NSD3 fragments used in this study. d, In vitro methylation reactions with rNucs and different concentrations of recombinant wild-type NSD3SET or NSD3SET-T1232A with 3H-SAM. Methylation intensity was measured by liquid scintillation counting. Error bars represent mean ± s.d. from three independent experiments. e, Western blot analysis with the indicated antibodies of whole cell extracts from NSD2-deficient HT1080 cells (to deplete H3K36me2 compared to control HT1080 cells (lanes 1 and 2)) overexpressing the indicated NSD3 variants. Total H3 was used as a loading control. f, Methylation assays as in Figure 2b with rNuc with linker DNA (187 bp) and increasing concentrations of enzyme as indicated. Western blot shows H3K36me2 generation, H3 is shown as a loading control. Bottom panel, Coomassie blue stain of NSD3 proteins.