Figure 8: Model for reprogramming of ClpAP by ClpS.
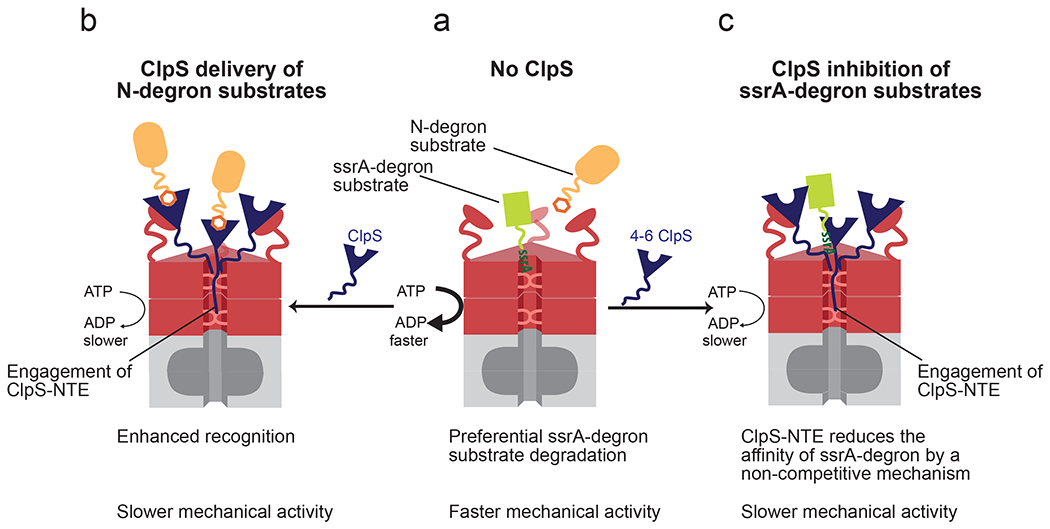
(a) In the absence of ClpS, ClpAP preferentially degrades ssrA-tagged substrates (green) relative to N-degron substrates (orange). (b) When the ClpS core binds the ClpA N-terminal domain, it positions the NTE for engagement by ClpA pore loops. ClpS NTE interactions with the translocation machinery suppress the rate of ATP hydrolysis by ClpA, slowing its mechanical activities. When N-degron substrates are present, ClpS also markedly enhances their recognition by ClpAP. (c) When ClpS and ssrA degron substrates are present, ClpS decreases the affinity for this substrate class by a non-competitive binding mechanism. Under these conditions the ClpS NTE also interacts with, and slows the translocation machinery of ClpA.
