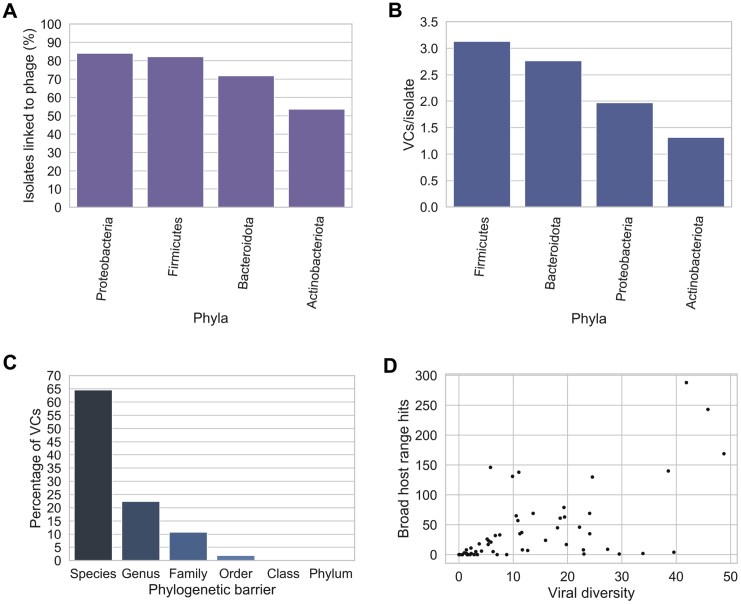
Figure S2.
Bacterial host assignment and host range for gut phage, related to Figure 2
A) Percentage of isolates of each phylum linked to phage by CRISPR spacers and prophage assignment. Actinobacteria had the lowest percentage of isolates predicted to be a phage host. Actinobacteria versus Bacteroidota (p = 0.007, test), Actinobacteria versus Proteobacteria (p = 0.0025, test), Actinobacteria versus Firmicutes (p = 1.01 × 10−5, test). B) The Firmicutes hosted the highest viral diversity (highest number of VCs/isolate). Firmicutes versus Bacteroidota (p = 0.021, test), Firmicutes versus Proteobacteria (p = 4.41 × 10-6, test), Firmicutes versus Actinobacteriota (p = 1.1 × 10−31, test) C) The majority of VCs were found to be restricted to infect a single species. However, a considerable number of VCs (~36%) had a broader host range (p = 0.0, binomial test). D) In general, the higher the viral diversity per bacterial genus, the higher the number of phages with broad host range (Spearman’s Rho = 0.6685, p = 3.91x10−9).