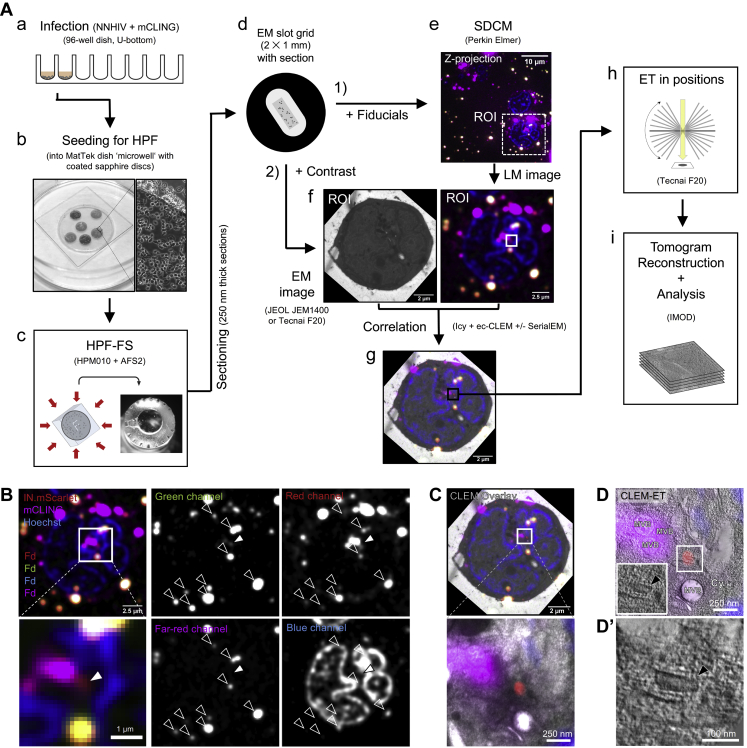
Figure S2.
Workflow for CLEM-ET visualization of HIV-1 post-entry complexes, related to Figures 2, 3, 4, and 5
(A) SupT1-R5 cells are incubated with IN.mScarlet-carrying NNHIV particles for 90 min at 16°C in a 96-well dish. Cells are stained with mCLING.Atto647N for an additional 10 min at 16°C and shifted to 37°C to initiate virus entry (a). For high pressure freezing, cells are transferred (at 37°C, in the presence of mCLING) to MatTek dish ‘microwell’ containing carbon- and retronectin-coated sapphire discs (b). At indicated times after temperature shift, cells adhered on sapphire discs are high pressure frozen, freeze substituted and embedded in Lowicryl resin (c). After polymerization, 250 nm-thick sections of the resin-embedded cell monolayer are transferred onto EM slot grids (d). Multichannel fluorescent fiducials (TetraSpeck microspheres) are applied to the grid and examined by spinning disc confocal microscopy (SDCM) (e). Regions of interests (ROI) are identified in the resulting z stacks (f, right) and sections on grids are contrasted for visualization by transmission electron microscopy (EM) (f, left). Using multi-fluorescent fiducials, which are visible in EM micrographs as dense 100 nm spheres; LM and EM images are correlated to identify positions of IN.mScarlet in the cell section (g). Finally, tilt series are acquired at the positions of the identified ROIs (h), tomograms are reconstructed and further analyzed (i). (B–D’) Identification and visualization of post-entry HIV-1 complexes in a representative resin section by 3D CLEM. (B) SDCM image of a 250-nm thick resin cell section. The fluorescence of IN.mScarlet (red) and mCLING.Atto647N (magenta) is retained after freeze substitution and resin embedding. Resin embedded sections are stained with Hoechst (blue) and decorated with multi-fluorescent fiducials for correlation (Fd; empty arrowheads). The white arrowheads indicate the position of the IN.mScarlet signal. (C) CLEM overlay of a fluorescence image with the respective electron micrograph (top) and enlarged detail corresponding to ROI where a tilt series was acquired (image below). (D–D’) Slices through tomographic reconstructions as overview correlated with SDCM image (D), and as rotated, enlarged tomographic slice (D’). The cone-shaped NNHIV capsid is observed in the cytosol (black arrowheads) at the position of mCLING-negative IN.mScarlet signal. Cy, cytosol; MVB, multivesicular body.