Figure S5.
Subtomogram averaging of hexamers and reconstruction of the capsid lattice, related to Figures 3 and 4
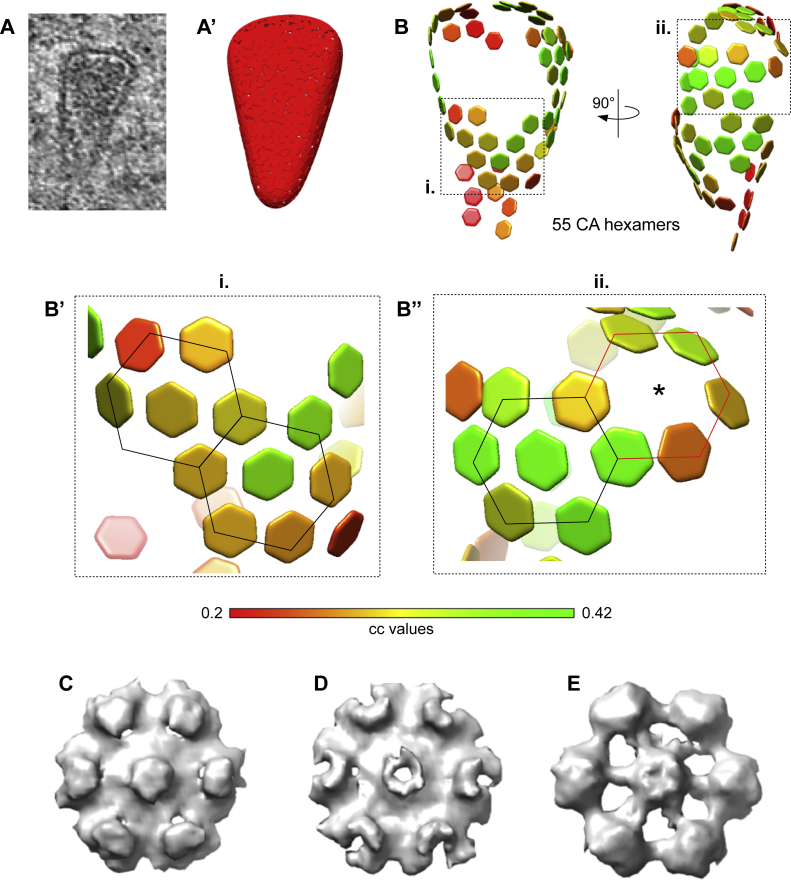
(A–B’’) Distribution of hexamers along the surface of a conical HIV-1 capsid before nuclear entry. (A) HIV-1 capsid in the cytosol of a cryo-FIB milled SupT1-R5 cell. Corresponding tomographic slice shown in Figure S3C and S4H. (A’) Segmented surface for the extraction of 1,137 initial subtomograms visualized in UCSF Chimera (Pettersen et al., 2004). The following procedure was applied on the identified structures to extract subtomograms for subtomogram averaging: (i) each capsid was manually segmented in IMOD, (ii) filtered volumes were generated from each segmentation in MATLAB and (iii) randomly distributed positions were extracted with approximately 4 × oversampling along the volume by defining their distance (based on the 10 nm hexamer-hexamer spacing) and the contour level of volume. (B) Lattice of 55 CA hexamers obtained after performing subtomogram averaging and cleaning the overlapping aligned subtomograms by cross-correlation (cc) values. cc values for each CA hexamer in the volume are shown color-coded as indicated in the colormap. (B’–B’’) The areas highlighted as i. and ii. of the lattice shown in (B), contain consecutive regular arrangements of six CA hexamers (highlighted by black lines) surrounding a seventh, central CA hexamer; in (B’’) six CA hexamers are localized to a highly curved region (red lines). The position of central CA hexamer was not detected (star). (C–E) Hexagonal subtomogram averages obtained from capsids in the cytosol (C) (conical capsid shown in panel A and in Figure S4G), within the NPC central channel (D) (conical capsid shown in Figure 3C), in the nucleus (E) (tubular capsid shown in Figure 4D).