Figure 1.
Clinical evaluation of SATB1 variants in neurodevelopmental disorders
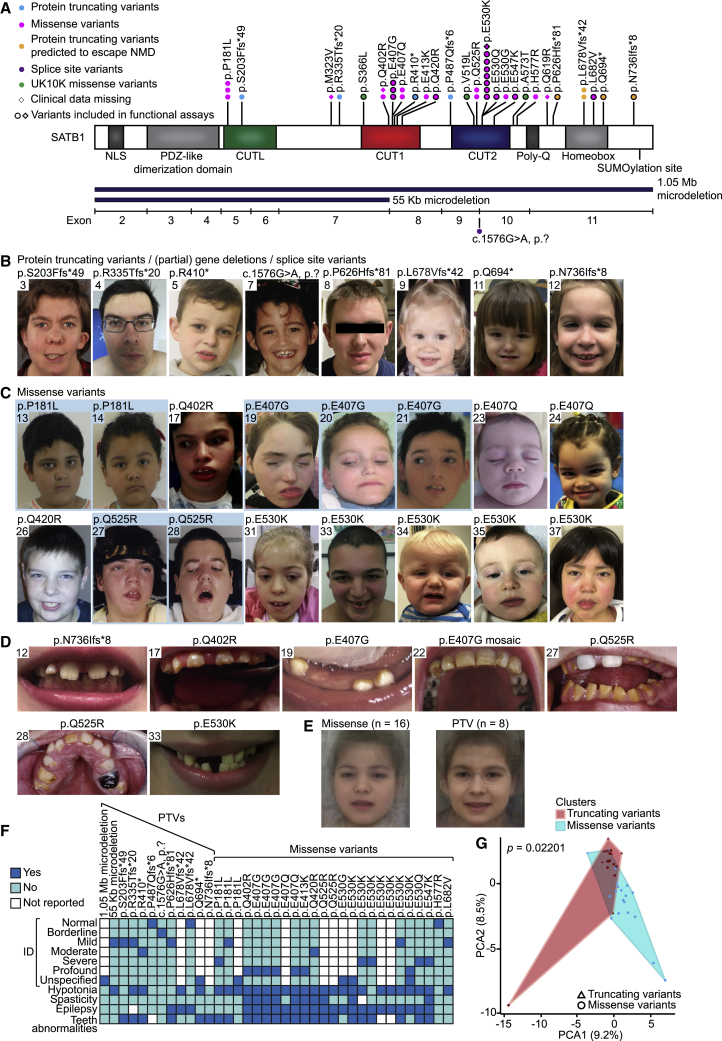
(A) Schematic representation of SATB1 (GenBank: NM_001131010.4/NP_001124482.1), including functional domains, with truncating variants labeled in cyan, truncating variants predicted to escape NMD in orange, splice site variants in purple, missense variants in magenta, and UK10K rare control missense variants in green. Deletions are shown in dark blue below the protein schematic, above a diagram showing the exon boundaries. We obtained clinical data for all individuals depicted by a circle.
(B and C) Facial photographs of individuals with (partial) gene deletions and truncations (B) and of individuals with missense variants (C). All depicted individuals show facial dysmorphisms and although overlapping features are seen, no consistent facial phenotype can be observed for the group as a whole. Overlapping facial dysmorphisms include facial asymmetry, high forehead, prominent ears, straight and/or full eyebrows, puffy eyelids, downslant of palpebral fissures, low nasal bridge, full nasal tip and full nasal alae, full lips with absent cupid’s bow, prominent cupid’s bow, or thin upper lip vermilion (Table S1B). Individuals with missense variants are more alike than individuals in the truncating cohorts, and we observed recognizable overlap between several individuals in the missense cohort (individuals 17, 27, 31, 37, the siblings 19, 20, and 21, and to a lesser extent individuals 24 and 35). A recognizable facial overlap between individuals with (partial) gene deletions and truncations could not be observed. Related individuals are marked with a blue box.
(D) Photographs of teeth abnormalities observed in individuals with SATB1 variants. Dental abnormalities are seen for all variant types and include widely spaced teeth, dental fragility, missing teeth, disorganized teeth positioning, and enamel discoloration (Table S1B).
(E) Computational average of facial photographs of 16 individuals with a missense variant (left) and 8 individuals with PTVs or (partial) gene deletions (right).
(F) Mosaic plot presenting a selection of clinical features.
(G) The Partitioning Around Medoids analysis of clustered HPO-standardized clinical data from 38 individuals with truncating (triangle) and missense (circle) variants shows a significant distinction between the clusters of individuals with missense variants (blue) and individuals with PTVs (red). Applying Bonferroni correction, a p value smaller than 0.025 was considered significant.
For analyses displayed in (F) and (G), individuals with absence of any clinical data and/or low-level mosaicism for the SATB1 variant were omitted (for details, see supplemental materials and methods).