Figure 2.
3D protein modeling of SATB1 missense variants in DNA-binding domains
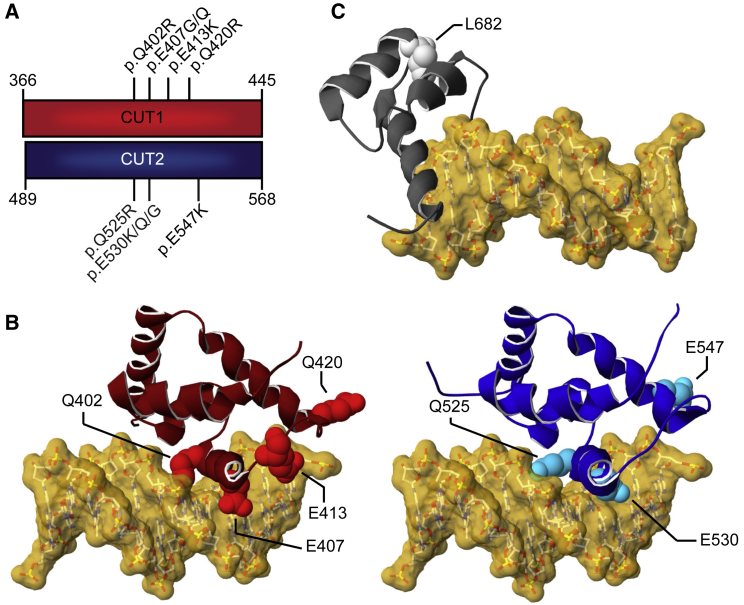
(A) Schematic representation of the aligned CUT1 and CUT2 DNA-binding domains. CUT1 and CUT2 domains have a high sequence identity (40%) and similarity (78%). Note that the recurrent p.Gln402Arg, p.Glu407Gly/p.Glu407Gln, and p.Gln525Arg, p.Glu530Gly/p.Glu530Lys/p.Glu530Gln variants affect equivalent positions within the respective CUT1 and CUT2 domains, while p.Gln420Arg in CUT1 and p.Glu547Lys in CUT2 affect cognate regions.
(B) 3D model of the SATB1 CUT1 domain (left; PDB: 2O4A) and CUT2 domain (right; based on PDB: 2CSF) in interaction with DNA (yellow). Mutated residues are highlighted in red for CUT1 and cyan for CUT2, along the ribbon visualization of the corresponding domains in burgundy and dark blue, respectively.
(C) 3D-homology model of the SATB1 homeobox domain (based on PDB: 1WI3 and 2D5V) in interaction with DNA (yellow). The mutated residue is shown in light gray along the ribbon visualization of the corresponding domain in dark gray.
(B and C) For more detailed descriptions of the different missense variants in our cohort, see Supplemental data.