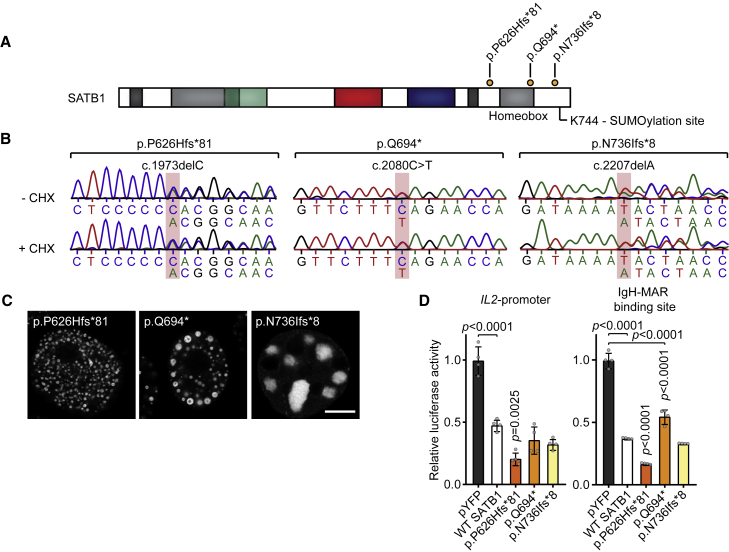
Figure 4.
SATB1 protein-truncating variants in the last exon escape NMD
(A) Schematic overview of the SATB1 protein, with truncating variants predicted to escape NMD that are included in functional assays labeled in orange. A potential SUMOylation site at position Lys744 is highlighted.
(B) Sanger sequencing traces of proband-derived EBV-immortalized lymphoblastoid cell lines treated with or without cycloheximide (CHX) to test for NMD. The mutated nucleotides are shaded in red. Transcripts from both alleles are present in both conditions showing that these variants escape NMD.
(C) Direct fluorescence super-resolution imaging of nuclei of HEK293T/17 cells expressing SATB1 truncating variants fused with a YFP tag. Scale bar = 5 μm. Compared to WT YFP-SATB1 (Figure 3), NMD-escaping variants show altered localization forming nuclear puncta or aggregates.
(D) Luciferase reporter assays using reporter constructs containing the IL2-promoter and the IgH matrix associated region (MAR) binding site. Values are expressed relative to the control (pYFP; black) and represent the mean ± SEM (n = 4, p values compared to WT SATB1 [white], one-way ANOVA and post hoc Bonferroni test). All NMD-escaping variants are transcriptionally active and show repression of the IL2-promoter and IgH-MAR binding site.